You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003957_01214
You are here: Home > Sequence: MGYG000003957_01214
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp014287585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp014287585 | |||||||||||
| CAZyme ID | MGYG000003957_01214 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2868; End: 5678 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 488 | 1.9e-70 | 0.4946808510638298 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.26e-24 | 24 | 547 | 9 | 509 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 5.06e-20 | 23 | 473 | 8 | 443 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 5.71e-20 | 22 | 473 | 36 | 470 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 7.94e-19 | 29 | 379 | 54 | 408 | beta-galactosidase. |
| pfam02837 | Glyco_hydro_2_N | 1.22e-14 | 29 | 220 | 3 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75482.1 | 0.0 | 5 | 936 | 3 | 934 |
| QRO25183.1 | 0.0 | 12 | 932 | 10 | 927 |
| SDS50045.1 | 0.0 | 14 | 936 | 14 | 948 |
| SCD20884.1 | 0.0 | 19 | 933 | 16 | 938 |
| ATP55310.1 | 0.0 | 28 | 932 | 31 | 948 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DYO_A | 2.07e-24 | 29 | 480 | 53 | 489 | ChainA, Beta-galactosidase [Escherichia coli K-12],3DYO_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYO_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYO_D Chain D, Beta-galactosidase [Escherichia coli K-12],3DYP_A Chain A, Beta-galactosidase [Escherichia coli K-12],3DYP_B Chain B, Beta-galactosidase [Escherichia coli K-12],3DYP_C Chain C, Beta-galactosidase [Escherichia coli K-12],3DYP_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
| 1F4A_A | 3.58e-24 | 29 | 480 | 51 | 487 | E.COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_B E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_C E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4A_D E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) [Escherichia coli],1F4H_A E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_B E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_C E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli],1F4H_D E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) [Escherichia coli] |
| 5A1A_A | 3.58e-24 | 29 | 480 | 52 | 488 | 2.2A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_B 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_C 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12],5A1A_D 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor [Escherichia coli K-12] |
| 3MUY_1 | 3.59e-24 | 29 | 480 | 53 | 489 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
| 3J7H_A | 3.59e-24 | 29 | 480 | 54 | 490 | Structureof beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_B Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_C Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],3J7H_D Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy [Escherichia coli K-12],4CKD_A Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_B Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_C Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],4CKD_D Model of complex between the E.coli enzyme beta-galactosidase and four single chain Fv antibody domains scFv13R4. [Escherichia coli K-12],6DRV_A Beta-galactosidase [Escherichia coli K-12],6DRV_B Beta-galactosidase [Escherichia coli K-12],6DRV_C Beta-galactosidase [Escherichia coli K-12],6DRV_D Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7ZWZ1 | 6.56e-24 | 29 | 480 | 54 | 490 | Beta-galactosidase OS=Escherichia coli O9:H4 (strain HS) OX=331112 GN=lacZ PE=3 SV=1 |
| A7ZI91 | 6.56e-24 | 29 | 480 | 54 | 490 | Beta-galactosidase OS=Escherichia coli O139:H28 (strain E24377A / ETEC) OX=331111 GN=lacZ PE=3 SV=1 |
| Q32JB6 | 1.49e-23 | 29 | 480 | 54 | 490 | Beta-galactosidase OS=Shigella dysenteriae serotype 1 (strain Sd197) OX=300267 GN=lacZ PE=3 SV=2 |
| P00722 | 1.96e-23 | 29 | 480 | 54 | 490 | Beta-galactosidase OS=Escherichia coli (strain K12) OX=83333 GN=lacZ PE=1 SV=2 |
| A7KGA5 | 1.96e-23 | 29 | 480 | 54 | 490 | Beta-galactosidase OS=Klebsiella pneumoniae OX=573 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
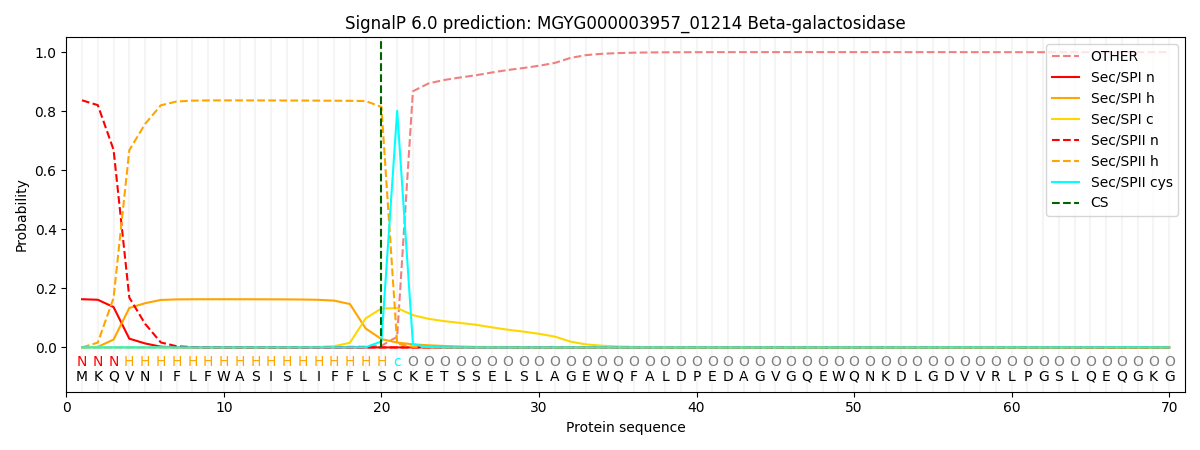
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000171 | 0.160478 | 0.839158 | 0.000058 | 0.000068 | 0.000062 |
