You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003958_01108
You are here: Home > Sequence: MGYG000003958_01108
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900545715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900545715 | |||||||||||
| CAZyme ID | MGYG000003958_01108 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 317026; End: 318618 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 92 | 480 | 1.2e-41 | 0.9570957095709571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.07e-32 | 168 | 480 | 15 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.01e-25 | 168 | 480 | 58 | 308 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.14e-21 | 168 | 486 | 81 | 343 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA28189.1 | 1.13e-125 | 45 | 530 | 28 | 504 |
| AHF92621.1 | 7.99e-121 | 42 | 520 | 15 | 481 |
| AVM47074.1 | 5.42e-120 | 34 | 520 | 7 | 486 |
| AWI10666.1 | 4.90e-119 | 45 | 524 | 1 | 474 |
| QQZ02681.1 | 3.99e-115 | 38 | 520 | 18 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHF_A | 5.62e-15 | 145 | 480 | 41 | 358 | Highlyactive enzymes by automated modular backbone assembly and sequence design [Escherichia coli] |
| 6FHE_A | 8.56e-14 | 173 | 478 | 73 | 325 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 6LPS_A | 3.03e-13 | 171 | 482 | 68 | 351 | ChainA, Beta-xylanase [Halalkalibacterium halodurans] |
| 2UWF_A | 1.00e-12 | 171 | 482 | 67 | 350 | ChainA, ALKALINE ACTIVE ENDOXYLANASE [Halalkalibacterium halodurans] |
| 5EBA_A | 1.33e-12 | 171 | 482 | 68 | 355 | Crystalstructure of aromatic mutant (Y343A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23556 | 1.20e-10 | 265 | 450 | 125 | 307 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=xynA PE=1 SV=1 |
| P40942 | 2.06e-10 | 173 | 482 | 105 | 381 | Thermostable celloxylanase OS=Thermoclostridium stercorarium OX=1510 GN=xynB PE=1 SV=1 |
| P07528 | 2.85e-10 | 171 | 482 | 113 | 396 | Endo-1,4-beta-xylanase A OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=xynA PE=1 SV=1 |
| O80596 | 8.56e-08 | 255 | 513 | 811 | 1035 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| P40943 | 3.53e-07 | 173 | 482 | 107 | 405 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
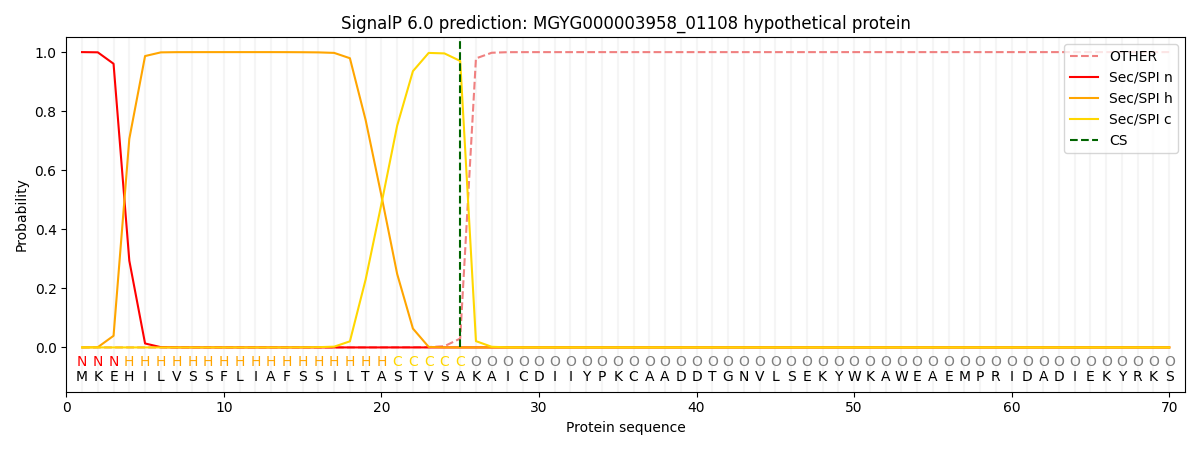
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000282 | 0.999096 | 0.000154 | 0.000157 | 0.000137 | 0.000127 |
