You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_00589
You are here: Home > Sequence: MGYG000004037_00589
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; | |||||||||||
| CAZyme ID | MGYG000004037_00589 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26202; End: 30116 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 833 | 1096 | 1.3e-74 | 0.8993055555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01095 | Pectinesterase | 1.38e-43 | 833 | 1143 | 2 | 298 | Pectinesterase. |
| PLN02773 | PLN02773 | 2.78e-38 | 835 | 1075 | 9 | 246 | pectinesterase |
| PLN02432 | PLN02432 | 3.74e-38 | 835 | 1145 | 15 | 291 | putative pectinesterase |
| PLN02990 | PLN02990 | 2.89e-37 | 833 | 1148 | 261 | 561 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02217 | PLN02217 | 1.02e-36 | 834 | 1148 | 253 | 551 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD39271.1 | 1.64e-305 | 99 | 1229 | 118 | 1247 |
| QCP72963.1 | 1.64e-305 | 99 | 1229 | 118 | 1247 |
| ANU62443.1 | 1.08e-282 | 200 | 1244 | 553 | 1596 |
| QQR10230.1 | 1.08e-282 | 200 | 1244 | 553 | 1596 |
| ASB37074.1 | 1.08e-282 | 200 | 1244 | 553 | 1596 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GQ8_A | 3.82e-31 | 834 | 1094 | 10 | 271 | Pectinmethylesterase from Carrot [Daucus carota] |
| 3UW0_A | 2.17e-28 | 828 | 1054 | 28 | 270 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 1XG2_A | 1.31e-27 | 833 | 1080 | 5 | 250 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 5C1E_A | 5.81e-24 | 841 | 1100 | 17 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 7.86e-24 | 841 | 1100 | 17 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q43043 | 7.36e-37 | 791 | 1090 | 6 | 318 | Pectinesterase OS=Petunia integrifolia OX=4103 GN=PPE1 PE=2 SV=1 |
| Q9FJ21 | 2.95e-32 | 834 | 1148 | 261 | 560 | Probable pectinesterase/pectinesterase inhibitor 58 OS=Arabidopsis thaliana OX=3702 GN=PME58 PE=2 SV=1 |
| Q8GXA1 | 3.81e-32 | 833 | 1157 | 258 | 567 | Probable pectinesterase/pectinesterase inhibitor 23 OS=Arabidopsis thaliana OX=3702 GN=PME23 PE=2 SV=3 |
| Q8GX86 | 3.82e-31 | 834 | 1148 | 257 | 555 | Probable pectinesterase/pectinesterase inhibitor 21 OS=Arabidopsis thaliana OX=3702 GN=PME21 PE=2 SV=2 |
| Q3E8Z8 | 5.47e-31 | 834 | 1148 | 254 | 552 | Putative pectinesterase/pectinesterase inhibitor 28 OS=Arabidopsis thaliana OX=3702 GN=PME28 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
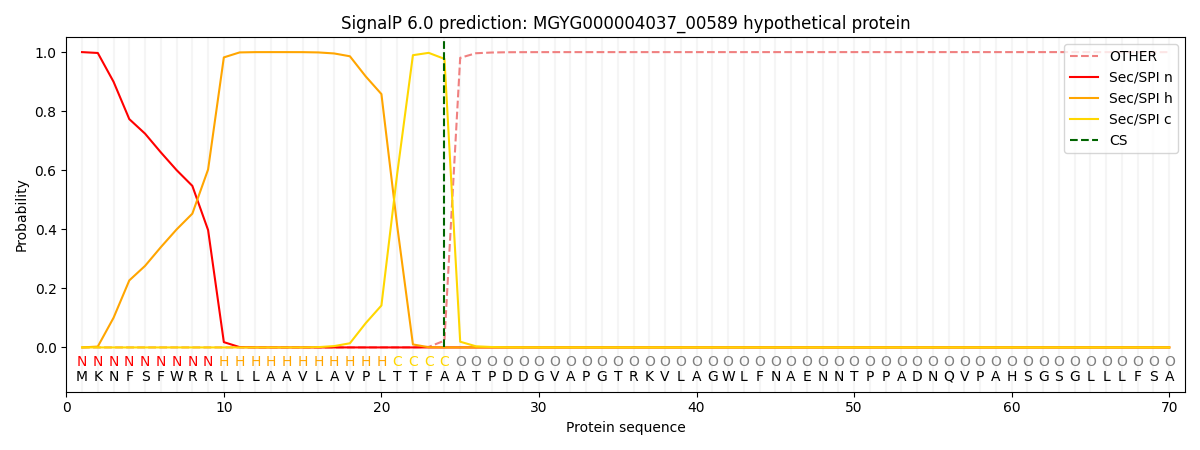
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000525 | 0.998471 | 0.000235 | 0.000283 | 0.000257 | 0.000200 |
