You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_01522
You are here: Home > Sequence: MGYG000004037_01522
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; | |||||||||||
| CAZyme ID | MGYG000004037_01522 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5580; End: 6476 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 3.10e-41 | 36 | 175 | 46 | 189 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| NF038016 | sporang_Gsm | 1.91e-30 | 48 | 174 | 175 | 312 | sporangiospore maturation cell wall hydrolase GsmA. The peptidoglycan-hydrolyzing enzyme GsmA occurs in some sporangia-forming members of the Actinobacteria, such as Actinoplanes missouriensis, and is required for proper separation of spores. GsmA proteins have one or two SH3 domains N-terminal to the hydrolase domain. |
| pfam01832 | Glucosaminidase | 3.59e-28 | 41 | 118 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| smart00047 | LYZ2 | 1.60e-27 | 33 | 174 | 8 | 147 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| PRK06347 | PRK06347 | 2.64e-20 | 55 | 284 | 172 | 426 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI64288.1 | 5.33e-113 | 23 | 298 | 20 | 298 |
| AHF12398.1 | 1.04e-110 | 24 | 298 | 19 | 299 |
| QUT71786.1 | 4.23e-108 | 1 | 298 | 1 | 301 |
| QQA10134.1 | 4.23e-108 | 1 | 298 | 1 | 301 |
| QDM08968.1 | 6.00e-108 | 10 | 298 | 7 | 301 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3FI7_A | 9.81e-13 | 24 | 174 | 20 | 183 | CrystalStructure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain [Listeria monocytogenes EGD-e] |
| 5T1Q_A | 9.96e-08 | 53 | 183 | 80 | 221 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O32083 | 5.27e-16 | 24 | 174 | 38 | 198 | Exo-glucosaminidase LytG OS=Bacillus subtilis (strain 168) OX=224308 GN=lytG PE=1 SV=1 |
| Q9X9J3 | 1.90e-13 | 24 | 164 | 147 | 301 | Peptidoglycan hydrolase FlgJ OS=Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633) OX=223926 GN=flgJ PE=3 SV=1 |
| Q9KQ15 | 3.21e-10 | 36 | 164 | 164 | 302 | Peptidoglycan hydrolase FlgJ OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=flgJ PE=3 SV=2 |
| Q2G222 | 3.15e-07 | 53 | 183 | 340 | 481 | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=SAOUHSC_02979 PE=1 SV=1 |
| P58231 | 6.32e-07 | 53 | 166 | 169 | 292 | Peptidoglycan hydrolase FlgJ OS=Escherichia coli O157:H7 OX=83334 GN=flgJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
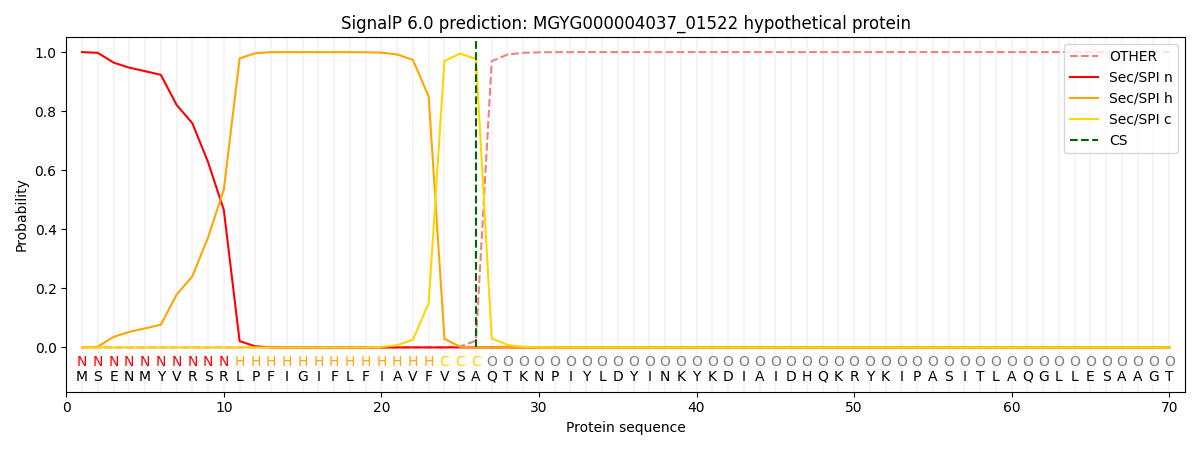
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000332 | 0.998995 | 0.000209 | 0.000145 | 0.000144 | 0.000136 |

