You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004042_01692
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485;
|
| CAZyme ID |
MGYG000004042_01692
|
| CAZy Family |
GH147 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004042 |
2392111 |
MAG |
United Kingdom |
Europe |
|
| Gene Location |
Start: 12230;
End: 14890
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH147 |
39 |
861 |
1.5e-284 |
0.9828431372549019 |
MGYG000004042_01692 has no CDD domain.
This protein is predicted as SP
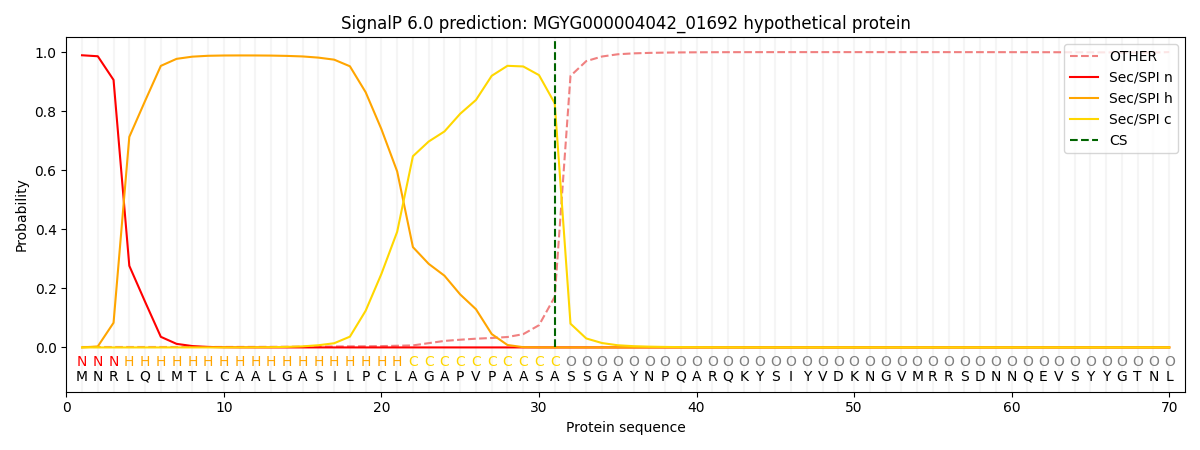
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.003434
|
0.984743
|
0.006857
|
0.004108
|
0.000516
|
0.000292
|
There is no transmembrane helices in MGYG000004042_01692.

