You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004043_01007
You are here: Home > Sequence: MGYG000004043_01007
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000004043_01007 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54666; End: 59174 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.29e-13 | 1051 | 1302 | 93 | 348 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam13205 | Big_5 | 2.49e-11 | 938 | 1034 | 1 | 106 | Bacterial Ig-like domain. |
| PLN02497 | PLN02497 | 3.17e-09 | 1187 | 1313 | 150 | 272 | probable pectinesterase |
| PLN02432 | PLN02432 | 1.21e-08 | 1166 | 1316 | 101 | 241 | putative pectinesterase |
| pfam18998 | Flg_new_2 | 1.47e-08 | 605 | 677 | 1 | 72 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD41145.1 | 0.0 | 1 | 1502 | 1 | 1495 |
| QCD38476.1 | 0.0 | 1 | 1501 | 1 | 1474 |
| QCP72166.1 | 0.0 | 1 | 1501 | 1 | 1474 |
| QNT65238.1 | 0.0 | 407 | 1489 | 24 | 1115 |
| QUT74167.1 | 0.0 | 3 | 1489 | 2 | 1417 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NTB_A | 5.01e-11 | 1106 | 1375 | 50 | 339 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 1QJV_A | 2.11e-10 | 1106 | 1375 | 50 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A9 | 3.27e-10 | 1106 | 1375 | 74 | 363 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| P0C1A8 | 1.81e-09 | 1106 | 1375 | 74 | 363 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
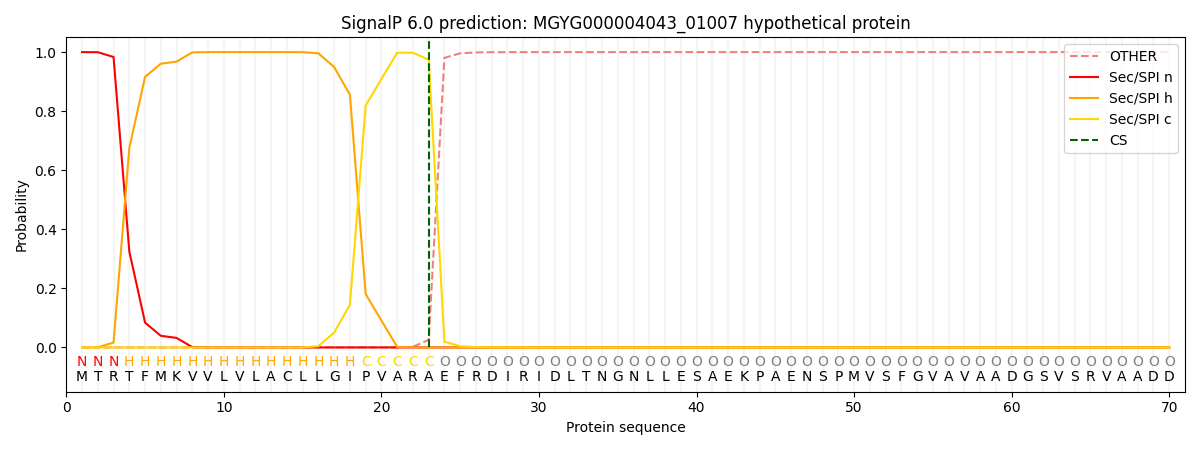
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000238 | 0.999090 | 0.000199 | 0.000157 | 0.000151 | 0.000138 |
