You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004076_00868
You are here: Home > Sequence: MGYG000004076_00868
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1623; | |||||||||||
| CAZyme ID | MGYG000004076_00868 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19291; End: 21573 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 63 | 743 | 2.6e-62 | 0.7220744680851063 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 7.54e-26 | 62 | 559 | 9 | 502 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.23e-14 | 68 | 476 | 15 | 422 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 1.25e-07 | 230 | 341 | 6 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 3.37e-07 | 148 | 471 | 141 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 3.50e-07 | 68 | 184 | 4 | 133 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV74706.1 | 2.42e-142 | 53 | 727 | 24 | 693 |
| QNN24094.1 | 4.20e-122 | 63 | 680 | 52 | 657 |
| QNN24101.1 | 8.05e-113 | 47 | 677 | 25 | 640 |
| QIJ31391.1 | 1.37e-108 | 63 | 688 | 40 | 665 |
| QBN20403.1 | 4.70e-108 | 68 | 688 | 32 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ECA_A | 6.69e-17 | 59 | 493 | 34 | 466 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 6U7I_A | 8.26e-17 | 63 | 471 | 10 | 412 | Faecalibacteriumprausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_B Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_C Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii],6U7I_D Faecalibacterium prausnitzii Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 7CWD_A | 4.54e-16 | 120 | 486 | 67 | 418 | ChainA, beta-glalactosidase [Niallia circulans],7CWI_A Chain A, beta-galactosidase [Niallia circulans] |
| 6JZ1_A | 1.03e-15 | 63 | 513 | 15 | 464 | Apostructure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ1_B Apo structure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ5_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ5_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ6_A b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ6_B b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_A b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_B b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ8_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus],6JZ8_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus] |
| 5Z18_A | 1.08e-15 | 63 | 513 | 39 | 488 | Thecrystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_B The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_C The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_D The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_E The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z18_F The crystal structure of Ruminococcus gnavus beta-glucuronidase [[Ruminococcus] gnavus],5Z19_A The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_B The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_C The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_D The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_E The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],5Z19_F The crystal structure of Ruminococcus gnavus beta-glucuronidase in complex with uronic isofagomine [[Ruminococcus] gnavus],6JZ2_A b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution [[Ruminococcus] gnavus],6JZ2_B b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution [[Ruminococcus] gnavus],6JZ3_A b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin [[Ruminococcus] gnavus],6JZ3_B b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin [[Ruminococcus] gnavus],6JZ4_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam [[Ruminococcus] gnavus],6JZ4_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 3.78e-15 | 120 | 475 | 109 | 436 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| P24131 | 1.24e-13 | 147 | 488 | 152 | 472 | Beta-galactosidase OS=Clostridium acetobutylicum OX=1488 GN=cbgA PE=2 SV=2 |
| Q9K9C6 | 2.04e-12 | 147 | 488 | 151 | 473 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| P23989 | 4.16e-11 | 147 | 489 | 151 | 476 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
| O52847 | 1.64e-10 | 147 | 473 | 164 | 483 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
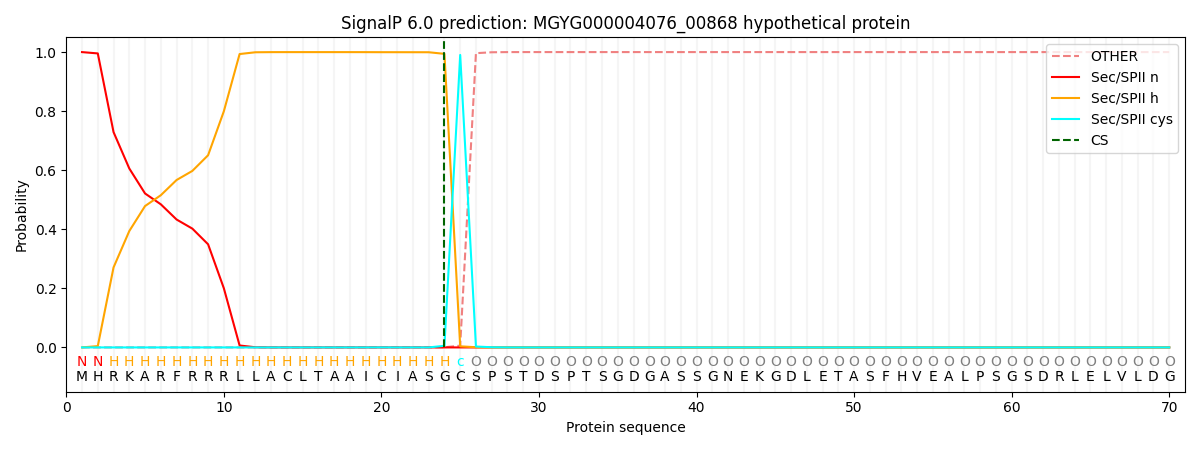
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000009 | 1.000052 | 0.000000 | 0.000000 | 0.000000 |
