You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004087_01763
You are here: Home > Sequence: MGYG000004087_01763
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterocloster sp900540675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster sp900540675 | |||||||||||
| CAZyme ID | MGYG000004087_01763 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29522; End: 34189 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 67 | 603 | 3.4e-154 | 0.9959266802443992 |
| CBM40 | 987 | 1150 | 7e-28 | 0.9217877094972067 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08428 | Rib | 2.90e-22 | 1268 | 1341 | 1 | 74 | Rib/alpha-like repeat. The region featured in this family is found repeated in a number of bacterial surface proteins, such as Rib and alpha. These are expressed by group B streptococci, and Rib is thought to confer protective immunity. |
| pfam02973 | Sialidase | 2.39e-13 | 996 | 1151 | 24 | 188 | Sialidase, N-terminal domain. |
| TIGR02331 | rib_alpha | 3.90e-13 | 1279 | 1341 | 2 | 66 | Rib/alpha/Esp surface antigen repeat. Sequences in this family are tandem repeats of about 79 amino acids, present in up to 14 copies in a protein and highly identical, even at the DNA level, within each protein. Sequences with these repeats are found in the Rib and alpha surface antigens of group B Streptococcus, Esp of Enterococcus faecalis, and related proteins of Lactobacillus. The repeat lacks Cys residues. Most members of this protein family also have the cell wall anchor motif LPXTG shared by many staphyloccal and streptococcal surface antigens. |
| COG5263 | COG5263 | 1.93e-10 | 1396 | 1551 | 197 | 312 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033840 | PspC_relate_1 | 7.41e-10 | 1408 | 1549 | 503 | 627 | PspC-related protein choline-binding protein 1. Members of this family share C-terminal homology to the choline-binding form of the pneumococcal surface antigen PspC, but not to its allelic LPXTG-anchored forms because they lack the choline-binding repeat region. Members of this family should not be confused with PspC itself, whose identity and function reflect regions N-terminal to the choline-binding region. See Iannelli, et al. (PMID: 11891047) for information about the different allelic forms of PspC. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT93619.1 | 7.64e-149 | 70 | 604 | 37 | 580 |
| ARV05068.1 | 4.10e-143 | 68 | 604 | 35 | 579 |
| ADO52688.1 | 4.27e-143 | 57 | 1027 | 23 | 1077 |
| BBA48469.1 | 1.12e-140 | 57 | 1027 | 23 | 1077 |
| BBA56588.1 | 2.89e-140 | 57 | 914 | 23 | 916 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 3.93e-157 | 66 | 603 | 7 | 576 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 7V6I_A | 1.08e-125 | 68 | 609 | 14 | 614 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 5.63e-125 | 70 | 478 | 247 | 644 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 1.25e-120 | 70 | 478 | 247 | 644 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 5GQC_A | 4.09e-120 | 68 | 606 | 18 | 599 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 4.01e-11 | 880 | 1111 | 461 | 669 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| Q27701 | 1.66e-10 | 969 | 1144 | 74 | 267 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| A0P8X0 | 4.68e-10 | 827 | 958 | 757 | 889 | Alpha-amylase OS=Niallia circulans OX=1397 GN=igtZ PE=1 SV=1 |
| P29767 | 5.67e-10 | 1019 | 1151 | 234 | 365 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P50899 | 7.63e-10 | 880 | 958 | 700 | 778 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
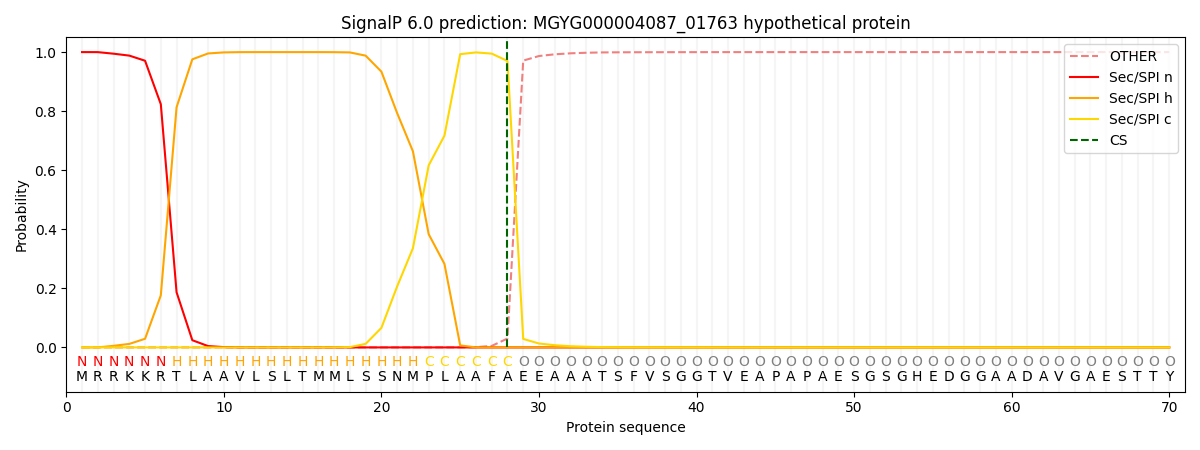
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.998922 | 0.000269 | 0.000197 | 0.000164 | 0.000145 |
