You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004109_00925
You are here: Home > Sequence: MGYG000004109_00925
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; ; ; ; | |||||||||||
| CAZyme ID | MGYG000004109_00925 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27154; End: 29118 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 177 | 459 | 2.8e-31 | 0.7476923076923077 |
| CBM66 | 496 | 631 | 2.2e-20 | 0.8838709677419355 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.29e-07 | 147 | 442 | 117 | 452 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02218 | PLN02218 | 0.002 | 229 | 456 | 197 | 410 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SBN61977.1 | 8.86e-107 | 24 | 477 | 47 | 480 |
| QKS86335.1 | 2.03e-99 | 20 | 477 | 36 | 475 |
| QGY47513.1 | 2.26e-87 | 39 | 476 | 60 | 471 |
| QIH36099.1 | 1.01e-42 | 40 | 476 | 57 | 471 |
| QQR16514.1 | 1.64e-42 | 46 | 476 | 31 | 464 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 2.26e-07 | 152 | 470 | 67 | 421 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
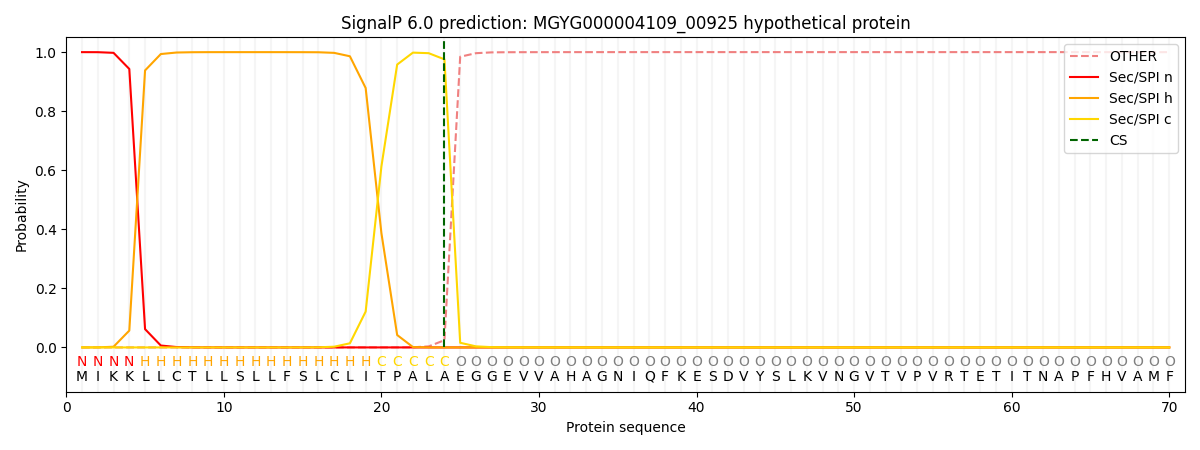
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000451 | 0.998028 | 0.000889 | 0.000226 | 0.000204 | 0.000181 |
