You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004132_01856
You are here: Home > Sequence: MGYG000004132_01856
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | V9D3004 sp900760345 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; V9D3004 sp900760345 | |||||||||||
| CAZyme ID | MGYG000004132_01856 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17920; End: 19590 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 181 | 548 | 5.1e-64 | 0.9504950495049505 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.45e-55 | 256 | 547 | 4 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.27e-53 | 171 | 549 | 4 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 6.89e-38 | 199 | 547 | 47 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| NF033186 | internalin_K | 8.96e-05 | 26 | 154 | 431 | 546 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
| PRK11633 | PRK11633 | 8.85e-04 | 103 | 164 | 89 | 151 | cell division protein DedD; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDM69886.1 | 2.54e-108 | 166 | 554 | 199 | 561 |
| ABX41884.1 | 2.67e-99 | 168 | 533 | 167 | 498 |
| BCZ47872.1 | 1.47e-96 | 167 | 550 | 42 | 399 |
| AFU34339.1 | 6.23e-96 | 170 | 550 | 173 | 519 |
| AGF55273.1 | 2.21e-93 | 168 | 550 | 40 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D5C_A | 3.06e-31 | 206 | 524 | 51 | 320 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 5OFJ_A | 1.62e-30 | 206 | 524 | 39 | 308 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 2W5F_A | 3.81e-30 | 169 | 556 | 181 | 535 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 5OFK_A | 1.05e-29 | 206 | 524 | 39 | 308 | Crystalstructure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose [Caldicellulosiruptor bescii DSM 6725],5OFL_A Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose [Caldicellulosiruptor bescii DSM 6725] |
| 2WYS_A | 8.09e-28 | 237 | 556 | 216 | 535 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23551 | 7.75e-80 | 170 | 533 | 37 | 366 | Endo-1,4-beta-xylanase A OS=Butyrivibrio fibrisolvens OX=831 GN=xynA PE=3 SV=1 |
| P29126 | 3.39e-35 | 204 | 533 | 654 | 936 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| P26223 | 1.04e-34 | 208 | 551 | 33 | 338 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| Q60037 | 4.11e-31 | 172 | 525 | 370 | 665 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 4.16e-30 | 165 | 525 | 359 | 661 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
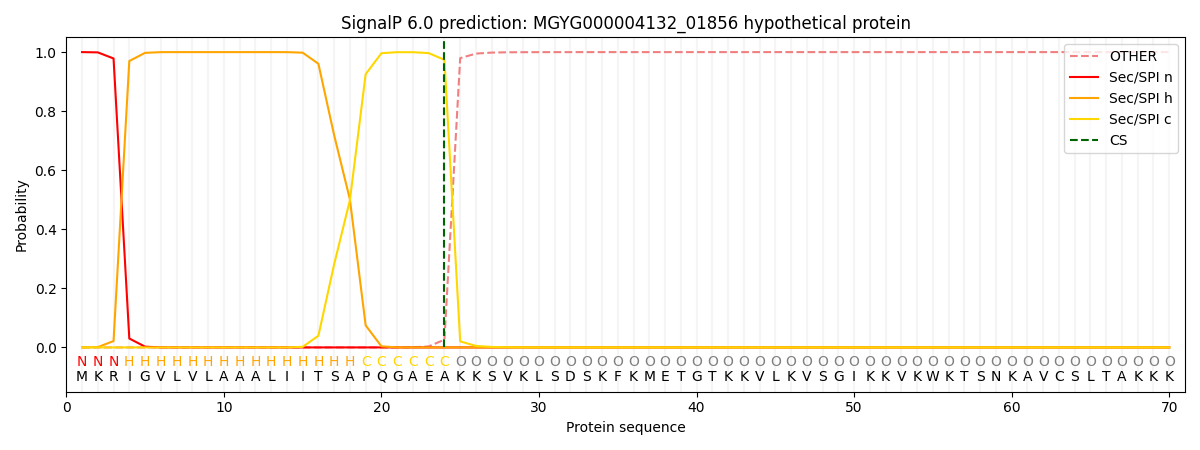
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000236 | 0.999168 | 0.000150 | 0.000153 | 0.000136 | 0.000125 |
