You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004151_02946
You are here: Home > Sequence: MGYG000004151_02946
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas citronellolis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas citronellolis | |||||||||||
| CAZyme ID | MGYG000004151_02946 | |||||||||||
| CAZy Family | CBM56 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45825; End: 47243 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3488 | COG3488 | 0.0 | 20 | 472 | 19 | 481 | Uncharacterized conserved protein with two CxxC motifs, DUF1111 family [General function prediction only]. |
| pfam06537 | DHOR | 1.90e-150 | 32 | 472 | 2 | 486 | Di-haem oxidoreductase, putative peroxidase. DHOR is a family of di-haem oxidoredictases. It carries the two characteristic Cys-X-Y-Cys-His haem-binding motifs. The C-terminal high-potential site functions as an electron transfer centre, and the N-terminal low-potential site corresponds to the peroxidatic centre. Its probable function is as a peroxidase. |
| COG1858 | MauG | 0.001 | 340 | 444 | 218 | 326 | Cytochrome c peroxidase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTD49369.1 | 1.46e-106 | 57 | 469 | 292 | 700 |
| ADO73975.1 | 7.13e-104 | 81 | 472 | 385 | 754 |
| QRK08295.1 | 4.55e-101 | 81 | 472 | 386 | 754 |
| QRN94766.1 | 1.98e-100 | 81 | 472 | 387 | 759 |
| AKJ01116.1 | 1.94e-99 | 81 | 472 | 369 | 740 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
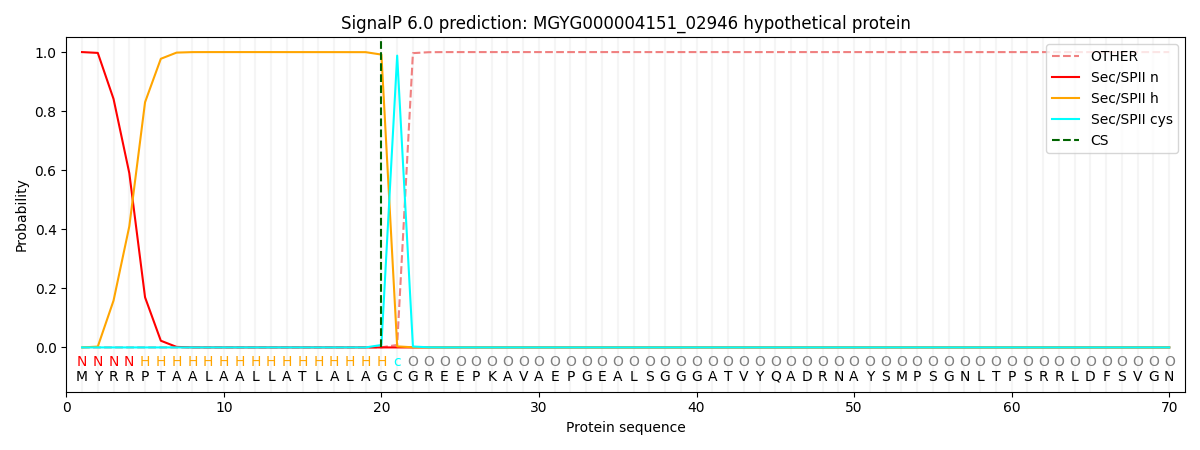
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000051 | 0.000000 | 0.000000 | 0.000000 |
