You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004157_00928
You are here: Home > Sequence: MGYG000004157_00928
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
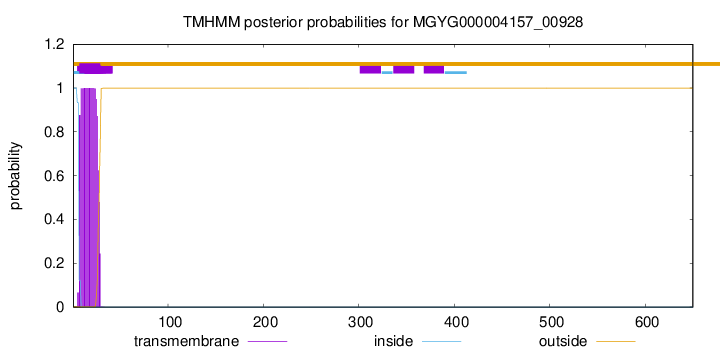
TMHMM annotations
Basic Information help
| Species | HGM11514 sp900757255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; HGM11514; HGM11514; HGM11514; HGM11514 sp900757255 | |||||||||||
| CAZyme ID | MGYG000004157_00928 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13240; End: 15192 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 429 | 647 | 1.9e-28 | 0.9782608695652174 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00413 | Glyco_hydrolase_16 | 5.44e-22 | 429 | 647 | 3 | 210 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| cd08023 | GH16_laminarinase_like | 1.60e-21 | 429 | 647 | 5 | 235 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| cd02178 | GH16_beta_agarase | 5.69e-15 | 420 | 647 | 18 | 257 | Beta-agarase, member of glycosyl hydrolase family 16. Beta-agarase is a glycosyl hydrolase family 16 (GH16) member that hydrolyzes the internal beta-1,4-linkage of agarose, a hydrophilic polysaccharide found in the cell wall of Rhodophyceaea, marine red algae. Agarose is a linear chain of galactose units linked by alternating L-alpha-1,3- and D-beta-1,4-linkages that are additionally modified by a 3,6-anhydro-bridge. Agarose forms thermo-reversible gels that are widely used in the food industry or as a laboratory medium. While beta-agarases are also found in two other families derived from the sequence-based classification of glycosyl hydrolases (GH50, and GH86) the GH16 members are most abundant. This domain adopts a curved beta-sandwich conformation, with a tunnel-shaped active site cavity, referred to as a jellyroll fold. |
| pfam00722 | Glyco_hydro_16 | 3.58e-12 | 458 | 645 | 5 | 168 | Glycosyl hydrolases family 16. |
| NF033190 | inl_like_NEAT_1 | 1.96e-10 | 19 | 202 | 557 | 745 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY47521.1 | 3.42e-47 | 433 | 649 | 42 | 258 |
| AUP79711.1 | 1.31e-44 | 449 | 650 | 61 | 259 |
| AWW29604.1 | 2.14e-44 | 436 | 649 | 57 | 265 |
| QNN21672.1 | 2.61e-37 | 421 | 648 | 43 | 256 |
| AXV49984.1 | 2.73e-34 | 432 | 649 | 42 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 1.27e-09 | 42 | 205 | 26 | 192 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 1.29e-09 | 42 | 205 | 5 | 171 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 3AXD_A | 3.85e-06 | 499 | 648 | 16 | 170 | Thetruncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form [Fibrobacter succinogenes subsp. succinogenes S85],3AXD_B The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form [Fibrobacter succinogenes subsp. succinogenes S85],3AXE_A The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) [Fibrobacter succinogenes subsp. succinogenes S85] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 4.01e-23 | 37 | 210 | 1677 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 2.95e-22 | 42 | 210 | 908 | 1082 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 7.23e-17 | 49 | 209 | 49 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P36917 | 7.90e-14 | 42 | 140 | 1056 | 1155 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P38537 | 4.40e-11 | 17 | 218 | 11 | 215 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
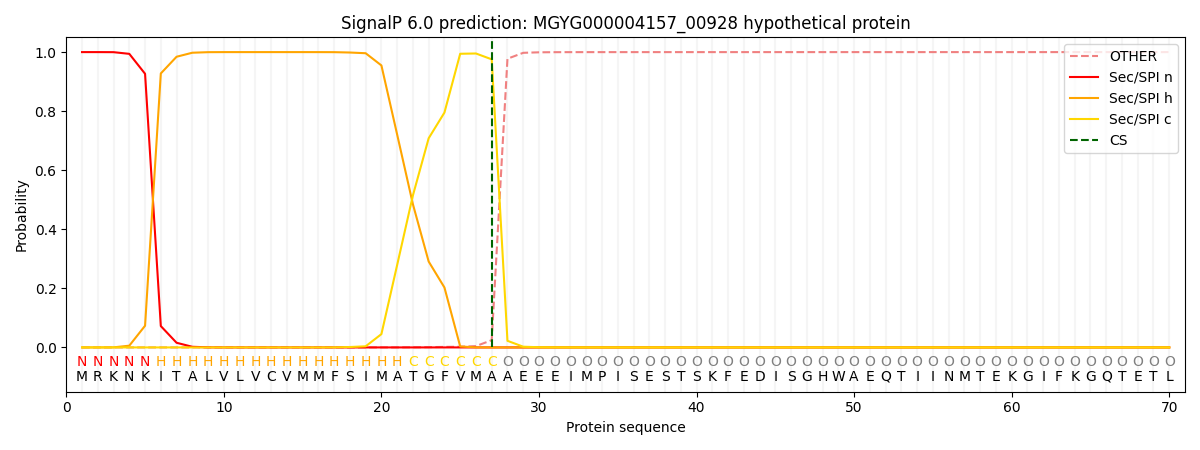
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000278 | 0.999018 | 0.000206 | 0.000170 | 0.000159 | 0.000145 |