You are browsing environment: HUMAN GUT
MGYG000004172_00141
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; ;
CAZyme ID
MGYG000004172_00141
CAZy Family
GT22
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000004172
3050906
MAG
United Kingdom
Europe
Gene Location
Start: 182305;
End: 183858
Strand: +
No EC number prediction in MGYG000004172_00141.
Family
Start
End
Evalue
family coverage
GT22
18
368
1.8e-44
0.8611825192802056
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam03901
Glyco_transf_22
9.36e-20
30
362
18
355
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
PLN02816
PLN02816
8.44e-12
29
327
57
347
mannosyltransferase
more
COG1807
ArnT
6.33e-07
172
368
158
367
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999948
0.000026
0.000005
0.000000
0.000000
0.000067
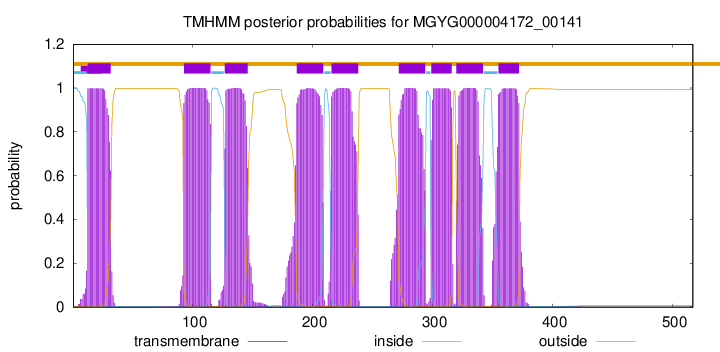
start
end
13
32
93
115
127
146
187
209
216
238
272
294
299
316
320
342
355
372