You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004188_01538
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides;
|
| CAZyme ID |
MGYG000004188_01538
|
| CAZy Family |
GH141 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004188 |
3351981 |
MAG |
United Kingdom |
Europe |
|
| Gene Location |
Start: 93160;
End: 95289
Strand: -
|
No EC number prediction in MGYG000004188_01538.
| Family |
Start |
End |
Evalue |
family coverage |
| GH141 |
27 |
301 |
3.7e-20 |
0.476280834914611 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
6.36e-04 |
309 |
496 |
3 |
136 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
0.004 |
457 |
578 |
2 |
112 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
This protein is predicted as SP
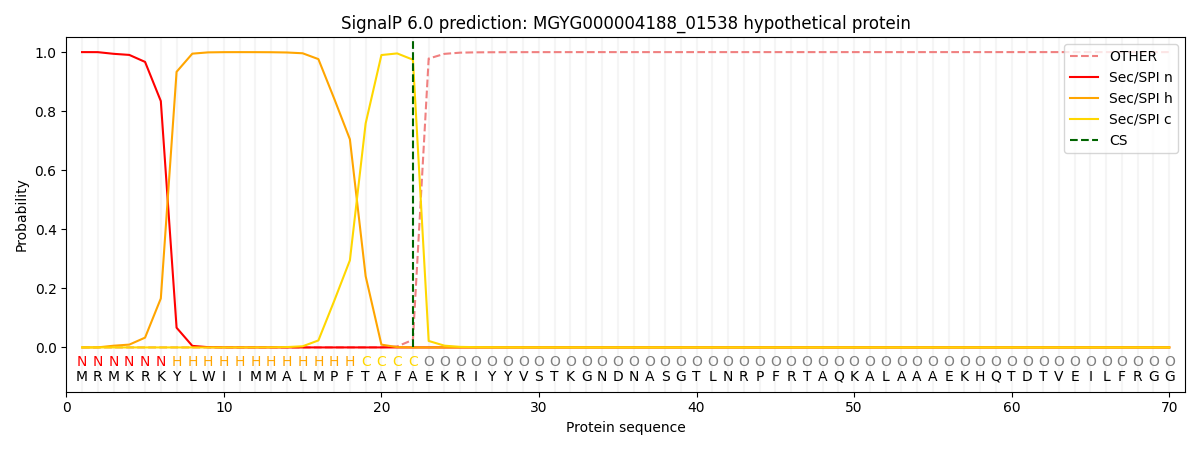
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000404
|
0.998729
|
0.000319
|
0.000185
|
0.000171
|
0.000161
|


