You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004197_01210
You are here: Home > Sequence: MGYG000004197_01210
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
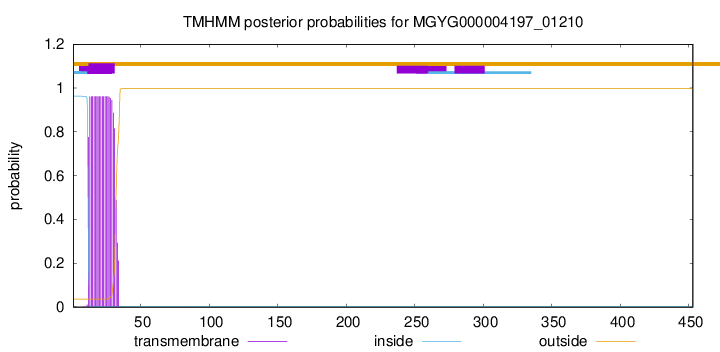
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; | |||||||||||
| CAZyme ID | MGYG000004197_01210 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3252; End: 4613 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 64 | 252 | 6.6e-20 | 0.45864661654135336 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.15e-40 | 63 | 419 | 1 | 342 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 2.43e-12 | 66 | 195 | 1 | 118 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK10206 | PRK10206 | 1.49e-06 | 124 | 222 | 51 | 145 | putative oxidoreductase; Provisional |
| pfam03435 | Sacchrp_dh_NADP | 7.64e-05 | 70 | 196 | 3 | 120 | Saccharopine dehydrogenase NADP binding domain. This family contains the NADP binding domain of saccharopine dehydrogenase. In some organisms this enzyme is found as a bifunctional polypeptide with lysine ketoglutarate reductase. The saccharopine dehydrogenase can also function as a saccharopine reductase. |
| COG1748 | Lys9 | 0.002 | 70 | 199 | 6 | 123 | Saccharopine dehydrogenase, NADP-dependent [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23801.1 | 4.30e-311 | 1 | 450 | 1 | 452 |
| BCG53919.1 | 1.68e-250 | 1 | 448 | 1 | 456 |
| BBK93797.1 | 1.14e-243 | 1 | 448 | 1 | 454 |
| QIX65768.1 | 1.14e-243 | 1 | 448 | 1 | 454 |
| QUT96603.1 | 1.14e-243 | 1 | 448 | 1 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CEA_A | 1.97e-15 | 64 | 355 | 7 | 263 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 4N54_A | 7.45e-09 | 64 | 280 | 13 | 205 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4MKX_A | 7.55e-09 | 64 | 280 | 16 | 208 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 3EC7_A | 3.11e-07 | 63 | 316 | 21 | 243 | CrystalStructure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_B Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_C Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_D Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_E Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_F Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_G Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],3EC7_H Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 1ZH8_A | 3.89e-07 | 128 | 370 | 71 | 309 | Crystalstructure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8],1ZH8_B Crystal structure of Oxidoreductase (TM0312) from Thermotoga maritima at 2.50 A resolution [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01S58 | 1.38e-10 | 5 | 252 | 2 | 229 | Glycosyl hydrolase family 109 protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6590 PE=3 SV=1 |
| Q9RK81 | 1.10e-09 | 1 | 226 | 1 | 214 | Glycosyl hydrolase family 109 protein OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO0529 PE=3 SV=1 |
| Q6M0B9 | 1.38e-08 | 66 | 386 | 2 | 284 | UDP-N-acetylglucosamine 3-dehydrogenase OS=Methanococcus maripaludis (strain S2 / LL) OX=267377 GN=MMP0352 PE=1 SV=1 |
| Q7MWF4 | 2.31e-07 | 43 | 242 | 37 | 241 | Glycosyl hydrolase family 109 protein OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=PG_0664 PE=3 SV=2 |
| A9N564 | 5.02e-07 | 66 | 316 | 3 | 222 | Inositol 2-dehydrogenase OS=Salmonella paratyphi B (strain ATCC BAA-1250 / SPB7) OX=1016998 GN=iolG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
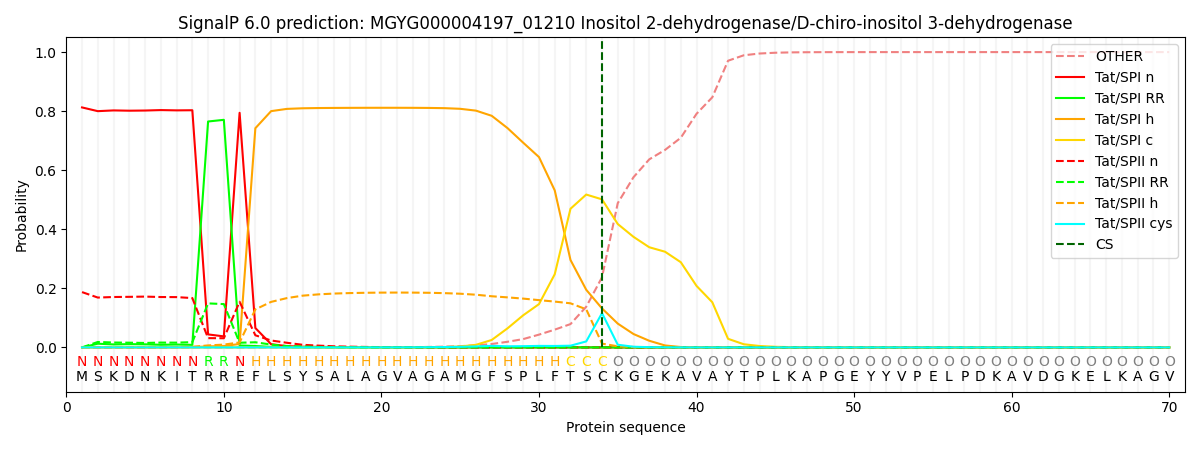
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000002 | 0.000001 | 0.000012 | 0.812646 | 0.187320 | 0.000000 |