You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004207_00275
You are here: Home > Sequence: MGYG000004207_00275
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA6398 sp003150315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA6398; UBA6398 sp003150315 | |||||||||||
| CAZyme ID | MGYG000004207_00275 | |||||||||||
| CAZy Family | GH127 | |||||||||||
| CAZyme Description | Non-reducing end beta-L-arabinofuranosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 58997; End: 60847 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH127 | 53 | 527 | 1.5e-62 | 0.933206106870229 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 2.54e-66 | 77 | 528 | 61 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 7.73e-51 | 71 | 528 | 58 | 501 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| cd04434 | LanC_like | 2.63e-04 | 86 | 217 | 64 | 192 | Cyclases involved in the biosynthesis of lantibiotics, and similar proteins. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthionine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as a precursor peptide and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans), in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. A related domain is also present in LanM and other pro- and eukaryotic proteins with poorly characterized functions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARE60508.1 | 4.46e-168 | 29 | 609 | 24 | 608 |
| AJQ29907.1 | 3.57e-43 | 72 | 560 | 74 | 593 |
| AJG99734.1 | 4.24e-40 | 72 | 541 | 74 | 572 |
| AQS05105.1 | 4.24e-40 | 72 | 541 | 74 | 572 |
| ADH61394.1 | 1.92e-39 | 99 | 551 | 99 | 573 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EX6_A | 1.76e-34 | 161 | 608 | 163 | 624 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 4QJY_A | 8.53e-32 | 78 | 546 | 81 | 569 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 5MQO_A | 4.59e-29 | 192 | 529 | 248 | 601 | Glycosidehydrolase BT_1003 [Bacteroides thetaiotaomicron] |
| 4QK0_A | 1.63e-28 | 78 | 546 | 81 | 569 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 7EXU_A | 7.50e-20 | 78 | 578 | 80 | 606 | ChainA, Non-reducing end beta-L-arabinofuranosidase [Bifidobacterium longum subsp. longum JCM 1217] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH8 | 5.33e-19 | 78 | 578 | 80 | 606 | Non-reducing end beta-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
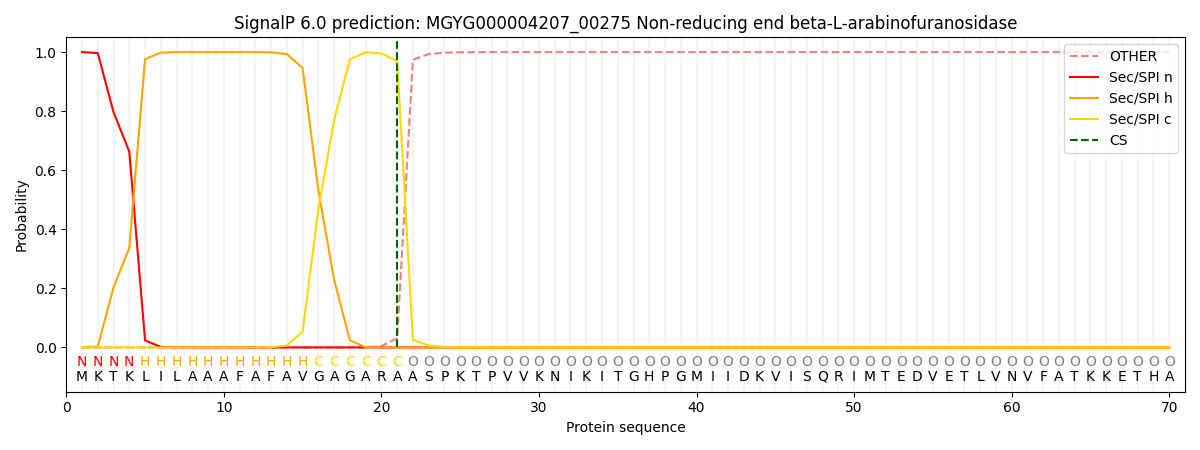
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000249 | 0.999101 | 0.000169 | 0.000160 | 0.000158 | 0.000140 |
