You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004219_02104
You are here: Home > Sequence: MGYG000004219_02104
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
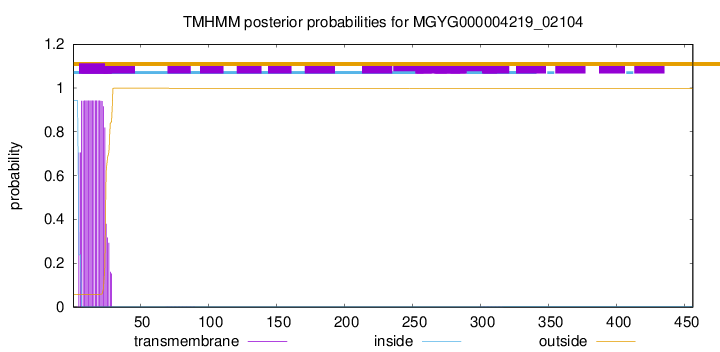
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; | |||||||||||
| CAZyme ID | MGYG000004219_02104 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10375; End: 11745 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 41 | 450 | 3.8e-119 | 0.9932735426008968 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01532 | Glyco_hydro_47 | 4.72e-135 | 41 | 450 | 1 | 451 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| PTZ00470 | PTZ00470 | 1.34e-110 | 35 | 449 | 73 | 515 | glycoside hydrolase family 47 protein; Provisional |
| COG1331 | YyaL | 5.18e-04 | 124 | 217 | 475 | 575 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIM36873.1 | 4.62e-253 | 3 | 455 | 2 | 450 |
| CDS95309.1 | 8.09e-251 | 30 | 455 | 33 | 457 |
| QQD11950.1 | 8.09e-251 | 30 | 455 | 33 | 457 |
| ANQ52185.2 | 2.39e-202 | 30 | 454 | 44 | 472 |
| QWG04411.1 | 2.39e-202 | 30 | 454 | 44 | 472 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5KKB_A | 1.58e-73 | 19 | 450 | 8 | 465 | Structureof mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus],5KKB_B Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex [Mus musculus] |
| 1NXC_A | 1.99e-73 | 19 | 450 | 6 | 463 | Structureof mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) [Mus musculus] |
| 4AYO_A | 2.90e-71 | 35 | 448 | 14 | 431 | Structureof The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 [Caulobacter sp. K31],4AYP_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with thiomannobioside [Caulobacter sp. K31],4AYQ_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with mannoimidazole [Caulobacter sp. K31],4AYR_A Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 in complex with noeuromycin [Caulobacter sp. K31],5MEH_A Crystal structure of alpha-1,2-mannosidase from Caulobacter K31 strain in complex with 1-deoxymannojirimycin [Caulobacter sp. K31],5NE5_A Crystal structure of family 47 alpha-1,2-mannosidase from Caulobacter K31 strain in complex with kifunensine [Caulobacter sp. K31] |
| 5KIJ_A | 2.58e-65 | 42 | 449 | 13 | 448 | Crystalstructure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
| 1FMI_A | 2.98e-65 | 42 | 449 | 18 | 453 | CrystalStructure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9C512 | 9.10e-75 | 36 | 449 | 99 | 533 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 OS=Arabidopsis thaliana OX=3702 GN=MNS1 PE=1 SV=1 |
| P45701 | 2.22e-73 | 25 | 448 | 3 | 452 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (Fragment) OS=Oryctolagus cuniculus OX=9986 GN=MAN1A1 PE=1 SV=1 |
| Q8H116 | 1.44e-71 | 36 | 449 | 100 | 534 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS2 OS=Arabidopsis thaliana OX=3702 GN=MNS2 PE=1 SV=1 |
| P45700 | 5.79e-71 | 19 | 450 | 183 | 640 | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA OS=Mus musculus OX=10090 GN=Man1a1 PE=1 SV=1 |
| Q9SXC9 | 8.10e-71 | 3 | 449 | 8 | 475 | Alpha-mannosidase I MNS5 OS=Arabidopsis thaliana OX=3702 GN=MNS5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
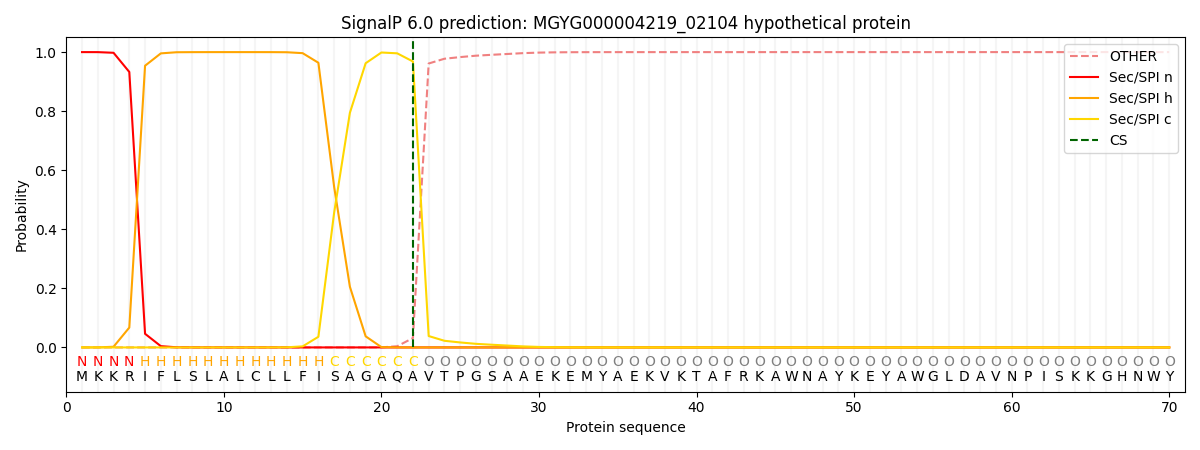
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000281 | 0.999034 | 0.000198 | 0.000168 | 0.000142 | 0.000137 |