You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004292_00326
You are here: Home > Sequence: MGYG000004292_00326
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1474 sp900552115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS1474; UMGS1474 sp900552115 | |||||||||||
| CAZyme ID | MGYG000004292_00326 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22257; End: 25970 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 682 | 999 | 3.9e-87 | 0.9017341040462428 |
| CBM32 | 1109 | 1228 | 6e-20 | 0.9274193548387096 |
| CBM6 | 570 | 676 | 2.5e-16 | 0.7318840579710145 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00812 | Alpha_L_fucos | 2.32e-88 | 678 | 1040 | 7 | 384 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| pfam01120 | Alpha_L_fucos | 1.06e-83 | 681 | 995 | 7 | 332 | Alpha-L-fucosidase. |
| COG3669 | AfuC | 1.40e-28 | 704 | 1034 | 1 | 374 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| cd04084 | CBM6_xylanase-like | 9.40e-21 | 578 | 675 | 28 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 2.60e-17 | 1106 | 1227 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASV73096.1 | 5.52e-110 | 682 | 1091 | 36 | 468 |
| AQQ71941.1 | 1.24e-105 | 682 | 1087 | 46 | 466 |
| QDV62750.1 | 2.26e-105 | 690 | 1089 | 31 | 447 |
| QNN24652.1 | 6.99e-105 | 682 | 1083 | 586 | 1007 |
| AQT69554.1 | 7.71e-105 | 678 | 1090 | 33 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GN6_A | 2.25e-71 | 683 | 1086 | 23 | 444 | Alpha-L-fucosidaseisoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_B Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_C Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_D Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_E Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus],6GN6_F Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus [Paenibacillus thiaminolyticus] |
| 7DB5_A | 8.72e-48 | 694 | 1050 | 37 | 413 | ChainA, Alpha-L-fucosidase [Vibrio sp. EJY3] |
| 6O1J_A | 4.55e-43 | 691 | 987 | 5 | 335 | ChainA, AlfC [Lacticaseibacillus casei],6O1J_B Chain B, AlfC [Lacticaseibacillus casei],6O1J_C Chain C, AlfC [Lacticaseibacillus casei],6O1J_D Chain D, AlfC [Lacticaseibacillus casei],6O1J_E Chain E, AlfC [Lacticaseibacillus casei],6O1J_F Chain F, AlfC [Lacticaseibacillus casei],6O1J_G Chain G, AlfC [Lacticaseibacillus casei],6O1J_H Chain H, AlfC [Lacticaseibacillus casei] |
| 6O18_A | 6.17e-43 | 691 | 987 | 5 | 335 | ChainA, AlfC [Lacticaseibacillus casei],6O18_B Chain B, AlfC [Lacticaseibacillus casei],6O18_C Chain C, AlfC [Lacticaseibacillus casei],6O18_D Chain D, AlfC [Lacticaseibacillus casei],6O1A_A Chain A, AlfC [Lacticaseibacillus casei],6O1A_B Chain B, AlfC [Lacticaseibacillus casei],6O1A_C Chain C, AlfC [Lacticaseibacillus casei],6O1A_D Chain D, AlfC [Lacticaseibacillus casei] |
| 6O1I_A | 2.09e-42 | 691 | 987 | 5 | 335 | ChainA, AlfC [Lacticaseibacillus casei],6O1I_B Chain B, AlfC [Lacticaseibacillus casei],6O1I_C Chain C, AlfC [Lacticaseibacillus casei],6O1I_D Chain D, AlfC [Lacticaseibacillus casei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10901 | 1.28e-38 | 693 | 1074 | 36 | 445 | Alpha-L-fucosidase OS=Dictyostelium discoideum OX=44689 GN=alfA PE=3 SV=1 |
| Q2KIM0 | 3.39e-35 | 692 | 1079 | 54 | 456 | Tissue alpha-L-fucosidase OS=Bos taurus OX=9913 GN=FUCA1 PE=2 SV=1 |
| Q99KR8 | 1.80e-34 | 677 | 1070 | 27 | 440 | Plasma alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca2 PE=1 SV=1 |
| Q9BTY2 | 1.99e-34 | 689 | 1051 | 45 | 426 | Plasma alpha-L-fucosidase OS=Homo sapiens OX=9606 GN=FUCA2 PE=1 SV=2 |
| P17164 | 3.32e-34 | 693 | 1061 | 48 | 431 | Tissue alpha-L-fucosidase OS=Rattus norvegicus OX=10116 GN=Fuca1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
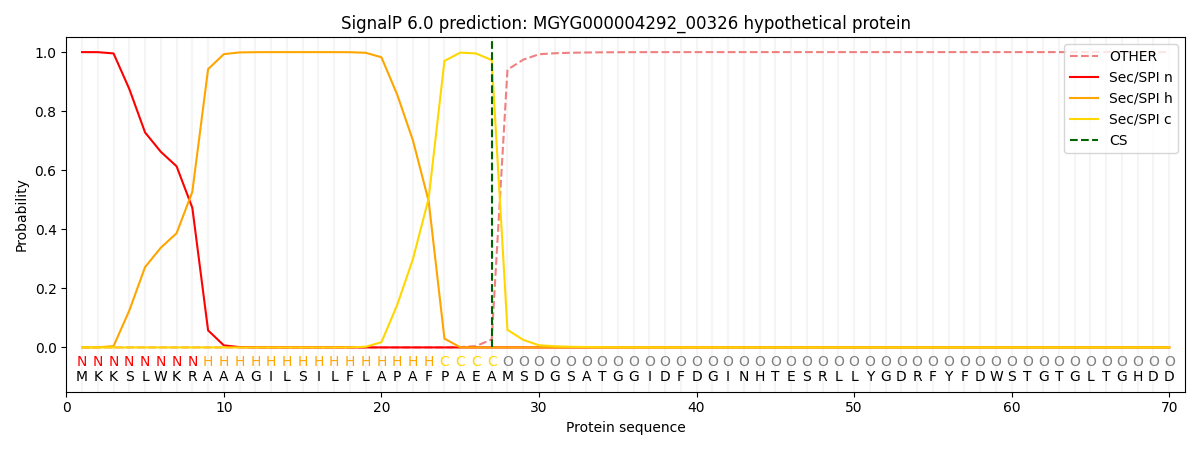
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000393 | 0.998604 | 0.000275 | 0.000257 | 0.000232 | 0.000212 |
