You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004296_01501
You are here: Home > Sequence: MGYG000004296_01501
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
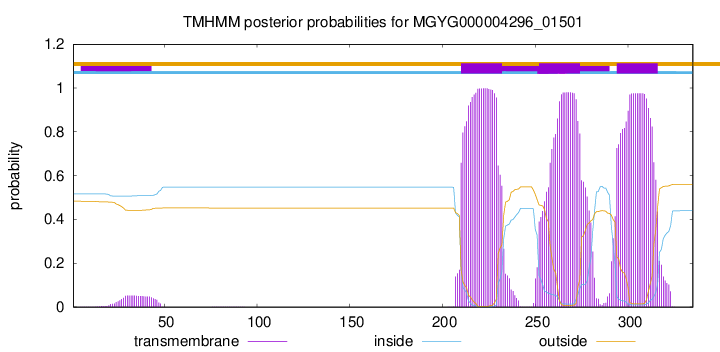
TMHMM annotations
Basic Information help
| Species | Ruminococcus_A faecicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Ruminococcus_A; Ruminococcus_A faecicola | |||||||||||
| CAZyme ID | MGYG000004296_01501 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 653; End: 1660 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 10 | 204 | 4.8e-58 | 0.8009259259259259 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03080 | PLN03080 | 1.09e-56 | 1 | 200 | 66 | 280 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 1.08e-46 | 3 | 204 | 47 | 234 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| cd10336 | SLC6sbd_Tyt1-Like | 4.35e-46 | 197 | 312 | 157 | 268 | solute carrier 6 subfamily, Fusobacterium nucleatum Tyt1-like; solute-binding domain. SLC6 proteins (also called the sodium- and chloride-dependent neurotransmitter transporter family or Na+/Cl--dependent transporter family) include neurotransmitter transporters (NTTs): these are sodium- and chloride-dependent plasma membrane transporters for the monoamine neurotransmitters serotonin (5-hydroxytryptamine), dopamine, and norepinephrine, and the amino acid neurotransmitters GABA and glycine. These NTTs are widely expressed in the mammalian brain, involved in regulating neurotransmitter signaling and homeostasis, and the target of a range of therapeutic drugs for the treatment of psychiatric diseases. Bacterial members of the SLC6 family include the LeuT amino acid transporter. An arrangement of 12 transmembrane (TM) helices appears to be as a common topological motif for eukaryotic and some prokaryotic and archaeal NTTs. However, this subfamily which contains the majority of bacterial members and some archaeal members, appears to contain only 11 TMs; for example the functional Fusobacterium nucleatum tyrosine transporter Tyt1. |
| pfam00933 | Glyco_hydro_3 | 5.24e-36 | 6 | 204 | 54 | 240 | Glycosyl hydrolase family 3 N terminal domain. |
| COG0733 | YocR | 1.01e-32 | 197 | 312 | 160 | 272 | Na+-dependent transporter, SNF family [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL10145.1 | 2.11e-249 | 1 | 335 | 1 | 335 |
| VCV24172.1 | 3.22e-246 | 1 | 330 | 1 | 330 |
| QNM03744.1 | 1.01e-128 | 1 | 200 | 22 | 221 |
| CBK91095.1 | 4.24e-127 | 1 | 200 | 22 | 221 |
| ACR74806.1 | 4.24e-127 | 1 | 200 | 22 | 221 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC7_A | 2.92e-46 | 8 | 204 | 46 | 252 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 2.92e-46 | 8 | 204 | 46 | 252 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 5A7M_A | 1.87e-36 | 9 | 197 | 74 | 268 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 5AE6_A | 1.88e-36 | 9 | 197 | 74 | 268 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 6Q7I_A | 1.22e-35 | 11 | 198 | 76 | 267 | GH3exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 2.31e-63 | 4 | 200 | 51 | 251 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q9SGZ5 | 4.43e-53 | 1 | 200 | 62 | 274 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| Q9LXD6 | 4.67e-53 | 11 | 204 | 83 | 284 | Beta-D-xylosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BXL3 PE=1 SV=1 |
| A1DJS5 | 3.16e-52 | 6 | 200 | 71 | 272 | Probable exo-1,4-beta-xylosidase xlnD OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xlnD PE=3 SV=1 |
| Q4WFI6 | 8.28e-52 | 6 | 200 | 71 | 272 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
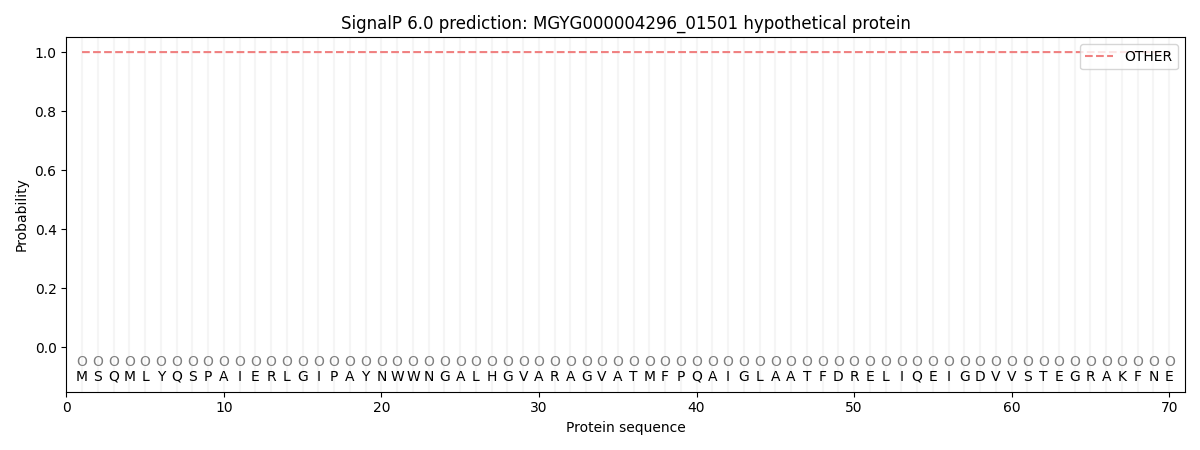
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000027 | 0.000008 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |