You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004345_01778
You are here: Home > Sequence: MGYG000004345_01778
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptococcus thermophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus thermophilus | |||||||||||
| CAZyme ID | MGYG000004345_01778 | |||||||||||
| CAZy Family | CBM41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4151; End: 4960 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02102 | pullulan_Gpos | 5.77e-109 | 1 | 262 | 224 | 482 | pullulanase, extracellular, Gram-positive. Pullulan is an unusual, industrially important polysaccharide in which short alpha-1,4 chains (maltotriose) are connected in alpha-1,6 linkages. Enzymes that cleave alpha-1,6 linkages in pullulan and release maltotriose are called pullulanases although pullulan itself may not be the natural substrate. In contrast, a glycogen debranching enzyme such GlgX, homologous to this family, can release glucose at alpha,1-6 linkages from glycogen first subjected to limit degradation by phosphorylase. Characterized members of this family include a surface-located pullulanase from Streptococcus pneumoniae () and an extracellular bifunctional amylase/pullulanase with C-terminal pullulanase activity (. |
| TIGR02104 | pulA_typeI | 2.00e-24 | 90 | 245 | 6 | 141 | pullulanase, type I. Pullulan is an unusual, industrially important polysaccharide in which short alpha-1,4 chains (maltotriose) are connected in alpha-1,6 linkages. Enzymes that cleave alpha-1,6 linkages in pullulan and release maltotriose are called pullulanases although pullulan itself may not be the natural substrate. This family consists of pullulanases related to the subfamilies described in TIGR02102 and TIGR02103 but having a different domain architecture with shorter sequences. Members are called type I pullulanases. |
| cd02860 | E_set_Pullulanase | 1.38e-22 | 94 | 189 | 1 | 88 | Early set domain associated with the catalytic domain of pullulanase (also called dextrinase and alpha-dextrin endo-1,6-alpha glucosidase). E or "early" set domains are associated with the catalytic domain of pullulanase at either the N-terminal or C-terminal end, and in a few instances at both ends. Pullulanase is an enzyme with activity similar to that of isoamylase; it cleaves 1,6-alpha-glucosidic linkages in pullulan, amylopectin, and glycogen, and in alpha-and beta-amylase limit-dextrins of amylopectin and glycogen. The E set domain of pullulanase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. |
| COG1523 | PulA | 3.60e-20 | 89 | 263 | 60 | 201 | Pullulanase/glycogen debranching enzyme [Carbohydrate transport and metabolism]. |
| PLN02877 | PLN02877 | 9.89e-15 | 89 | 260 | 208 | 369 | alpha-amylase/limit dextrinase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYK22274.1 | 3.71e-194 | 1 | 269 | 1 | 269 |
| CAD0169631.1 | 3.71e-194 | 1 | 269 | 1 | 269 |
| CAD0167385.1 | 3.71e-194 | 1 | 269 | 1 | 269 |
| SSC62485.1 | 3.71e-194 | 1 | 269 | 1 | 269 |
| CAD0124049.1 | 3.71e-194 | 1 | 269 | 1 | 269 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YA1_A | 1.18e-85 | 1 | 262 | 227 | 489 | Productcomplex of a multi-modular glycogen-degrading pneumococcal virulence factor SpuA [Streptococcus pneumoniae TIGR4] |
| 3FAW_A | 3.11e-77 | 4 | 262 | 43 | 298 | CrystalStructure of the Group B Streptococcus Pullulanase SAP [Streptococcus agalactiae COH1],3FAX_A The crystal structure of GBS pullulanase SAP in complex with maltotetraose [Streptococcus agalactiae COH1] |
| 2YA2_A | 1.28e-67 | 82 | 262 | 2 | 183 | CatalyticModule of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. [Streptococcus pneumoniae TIGR4] |
| 2YA0_A | 1.40e-67 | 82 | 262 | 1 | 182 | CatalyticModule of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA [Streptococcus pneumoniae TIGR4] |
| 6JEQ_A | 5.88e-13 | 89 | 243 | 33 | 167 | Crystalstructure of Pullulanase from Paenibacillus barengoltzii complex with beta-cyclodextrin [Paenibacillus barengoltzii],6JFJ_A Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin [Paenibacillus barengoltzii],6JFX_A Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltopentaose [Paenibacillus barengoltzii],6JHF_A Crystal structure of apo Pullulanase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],6JHG_A Crystal structure of apo Pullulanase from Paenibacillus barengoltzii in space group P212121 [Paenibacillus barengoltzii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9F930 | 3.14e-84 | 1 | 262 | 363 | 625 | Pullulanase A OS=Streptococcus pneumoniae OX=1313 GN=spuA PE=1 SV=1 |
| A0A0H2ZL64 | 3.67e-84 | 1 | 262 | 341 | 603 | Pullulanase A OS=Streptococcus pneumoniae serotype 2 (strain D39 / NCTC 7466) OX=373153 GN=spuA PE=3 SV=1 |
| A0A0H2UNG0 | 5.73e-84 | 1 | 262 | 356 | 618 | Pullulanase A OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=spuA PE=1 SV=1 |
| O33840 | 2.93e-10 | 4 | 243 | 129 | 352 | Pullulanase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pulA PE=1 SV=2 |
| Q8GTR4 | 9.84e-10 | 89 | 251 | 201 | 353 | Pullulanase 1, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=PU1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
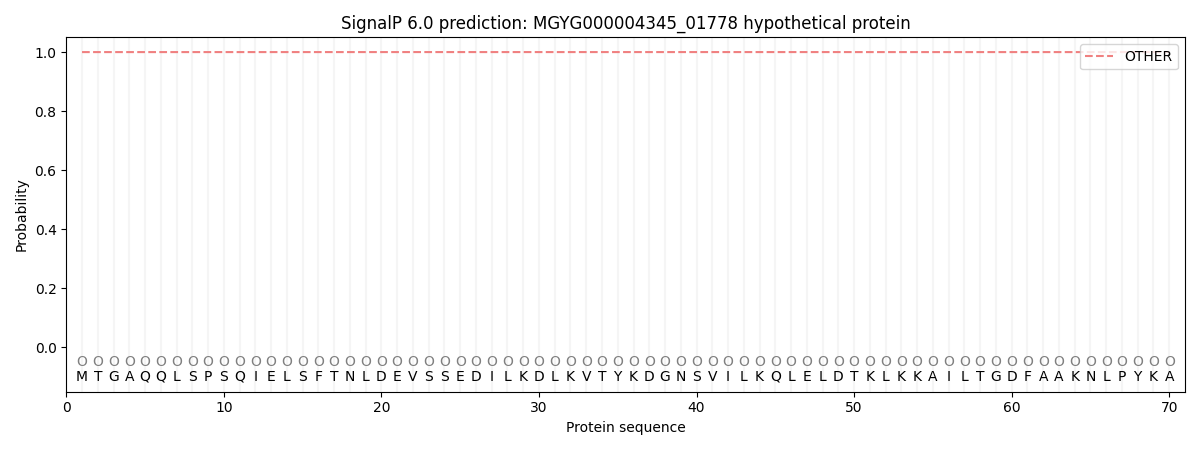
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000065 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
