You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004369_00945
You are here: Home > Sequence: MGYG000004369_00945
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
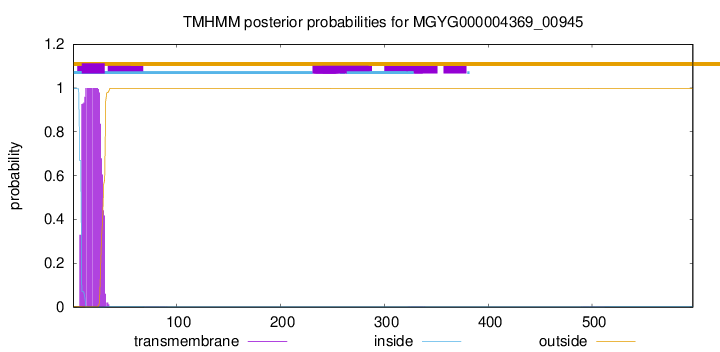
TMHMM annotations
Basic Information help
| Species | CAG-882 sp000435595 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-882; CAG-882 sp000435595 | |||||||||||
| CAZyme ID | MGYG000004369_00945 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35294; End: 37087 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 83 | 369 | 2.6e-86 | 0.9855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.86e-36 | 66 | 365 | 1 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.43e-21 | 14 | 359 | 18 | 352 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO05195.1 | 4.48e-220 | 39 | 595 | 43 | 593 |
| CBK96866.1 | 6.47e-202 | 39 | 593 | 31 | 581 |
| VEU81114.1 | 4.45e-193 | 46 | 593 | 31 | 575 |
| QTE66996.1 | 6.80e-175 | 49 | 597 | 31 | 592 |
| AHF25528.1 | 1.34e-173 | 49 | 594 | 16 | 574 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZMR_A | 6.74e-51 | 39 | 378 | 106 | 450 | Bacteroidesovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus],3ZMR_B Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus] |
| 6XRK_A | 1.83e-50 | 48 | 386 | 23 | 363 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 6WQP_A | 1.52e-48 | 46 | 381 | 13 | 343 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6MQ4_A | 7.61e-48 | 46 | 396 | 9 | 352 | ChainA, cellulase [Acetivibrio cellulolyticus] |
| 5D9N_A | 1.99e-46 | 57 | 347 | 21 | 295 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXT7 | 6.31e-50 | 39 | 378 | 141 | 485 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| O08342 | 6.75e-48 | 31 | 369 | 22 | 367 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P23660 | 7.68e-48 | 41 | 386 | 19 | 355 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28621 | 3.96e-45 | 29 | 386 | 23 | 366 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P54937 | 1.11e-43 | 51 | 404 | 41 | 389 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
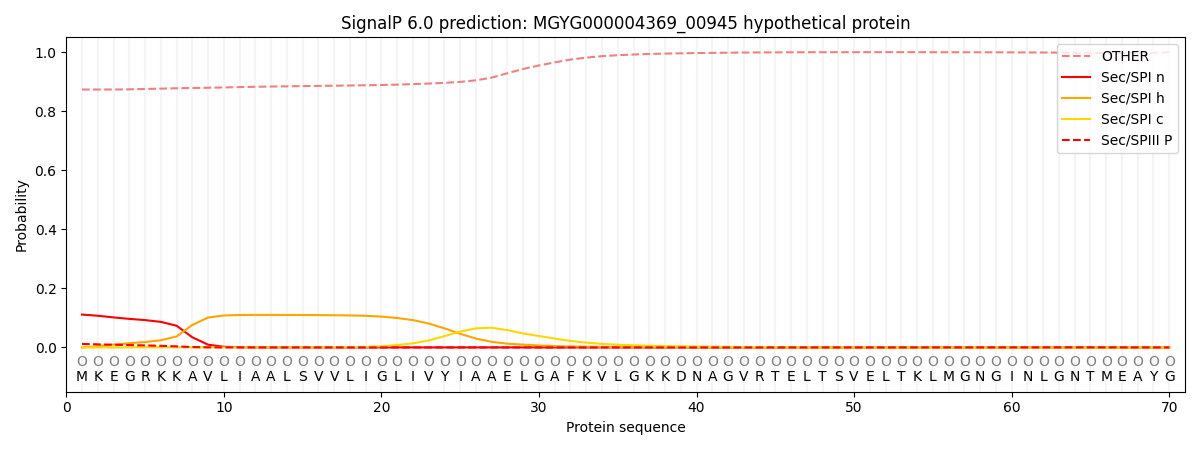
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.878575 | 0.102804 | 0.004631 | 0.000660 | 0.000394 | 0.012958 |