You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004469_01474
You are here: Home > Sequence: MGYG000004469_01474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900545315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900545315 | |||||||||||
| CAZyme ID | MGYG000004469_01474 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | Cytoplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 40991; End: 42496 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 47 | 471 | 5.6e-58 | 0.8553971486761711 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01204 | Trehalase | 2.74e-44 | 103 | 441 | 114 | 465 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| COG1626 | TreA | 6.75e-33 | 103 | 439 | 155 | 507 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PLN02567 | PLN02567 | 5.34e-31 | 97 | 338 | 127 | 383 | alpha,alpha-trehalase |
| PRK13271 | treA | 1.02e-21 | 103 | 439 | 146 | 492 | alpha,alpha-trehalase TreA. |
| PRK13272 | treA | 1.16e-21 | 75 | 435 | 110 | 488 | alpha,alpha-trehalase TreA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG53869.1 | 1.64e-172 | 69 | 499 | 22 | 452 |
| BCG53868.1 | 8.93e-160 | 74 | 499 | 15 | 440 |
| AHM60760.1 | 3.15e-93 | 75 | 487 | 26 | 440 |
| QQR16510.1 | 6.71e-70 | 103 | 496 | 55 | 429 |
| ANU58582.1 | 6.71e-70 | 103 | 496 | 55 | 429 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M4A_A | 8.24e-21 | 107 | 441 | 146 | 519 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae in complex with trehalose [Saccharomyces cerevisiae] |
| 5NIS_A | 9.16e-21 | 107 | 441 | 199 | 572 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae S288C] |
| 5JTA_A | 1.04e-20 | 107 | 441 | 294 | 667 | Neutraltrehalase Nth1 from Saccharomyces cerevisiae [Saccharomyces cerevisiae] |
| 5N6N_C | 1.05e-20 | 107 | 441 | 299 | 672 | CRYSTALSTRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX [Saccharomyces cerevisiae S288C] |
| 7E9X_A | 1.06e-12 | 82 | 338 | 129 | 396 | ChainA, Trehalase [Arabidopsis thaliana],7E9X_B Chain B, Trehalase [Arabidopsis thaliana],7E9X_C Chain C, Trehalase [Arabidopsis thaliana],7E9X_D Chain D, Trehalase [Arabidopsis thaliana],7EAW_A Chain A, Trehalase [Arabidopsis thaliana],7EAW_B Chain B, Trehalase [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49381 | 4.42e-23 | 107 | 441 | 297 | 669 | Cytosolic neutral trehalase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=NTH1 PE=1 SV=1 |
| P35172 | 1.08e-22 | 107 | 441 | 323 | 696 | Probable trehalase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=NTH2 PE=1 SV=1 |
| O42783 | 5.76e-21 | 114 | 441 | 288 | 658 | Cytosolic neutral trehalase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=treB PE=2 SV=2 |
| P32356 | 5.72e-20 | 107 | 441 | 294 | 667 | Cytosolic neutral trehalase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=NTH1 PE=1 SV=3 |
| P59765 | 9.81e-20 | 103 | 439 | 147 | 495 | Putative periplasmic trehalase OS=Salmonella typhi OX=90370 GN=treA PE=5 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
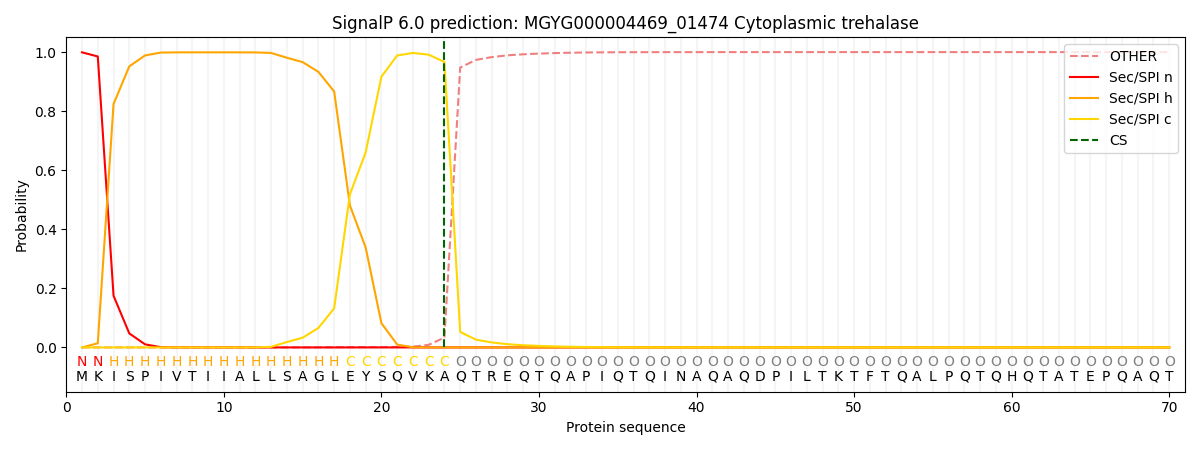
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000336 | 0.997609 | 0.001511 | 0.000196 | 0.000186 | 0.000151 |
