You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004483_01343
You are here: Home > Sequence: MGYG000004483_01343
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-873 sp900554715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900554715 | |||||||||||
| CAZyme ID | MGYG000004483_01343 | |||||||||||
| CAZy Family | GH171 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25988; End: 27154 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH171 | 43 | 386 | 1.7e-111 | 0.9971830985915493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07075 | DUF1343 | 5.55e-169 | 44 | 386 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| COG3876 | YbbC | 2.24e-87 | 17 | 386 | 19 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| cd01967 | Nitrogenase_MoFe_alpha_like | 0.003 | 28 | 109 | 274 | 355 | Nitrogenase_MoFe_alpha_like: Nitrogenase MoFe protein, alpha subunit_like. The nitrogenase enzyme catalyzes the ATP-dependent reduction of dinitrogen to ammonia. Three genetically distinct types of nitrogenase systems are known to exist: a molybdenum-dependent nitrogenase (Mo-nitrogenase), a vanadium dependent nitrogenase (V-nitrogenase), and an iron-only nitrogenase (Fe-nitrogenase). These nitrogenase systems consist of component 1 (MoFe protein, VFe protein or, FeFe protein respectively) and, component 2 (Fe protein). This group contains the alpha subunit of component 1 of all three different forms. The most widespread and best characterized of these systems is the Mo-nitrogenase. MoFe is an alpha2beta2 tetramer, the alternative nitrogenases are alpha2beta2delta2 hexamers having alpha and beta subunits similar to the alpha and beta subunits of MoFe. The role of the delta subunit is unknown. For MoFe, each alphabeta pair of subunits contains one P-cluster (located at the alphabeta interface) and, one molecule of iron molybdenum cofactor (FeMoco) contained within the alpha subunit. The Fe protein is a homodimer which contains, a single [4Fe-4S] cluster from which electrons are transferred to the P-cluster of the MoFe and in turn, to FeMoCo the site of substrate reduction. The V-nitrogenase requires an iron-vanadium cofactor (FeVco), the iron only-nitrogenase an iron only cofactor (FeFeco). These cofactors are analogous to the FeMoco. The V-nitrogenase has P clusters identical to those of MoFe. In addition to N2, nitrogenase also catalyzes the reduction of a variety of other substrates such as acetylene The V-nitrogenase differs from the Mo- nitrogenase in that it produces free hydrazine, as a minor product during dinitrogen reduction and, ethane as a minor product during acetylene reduction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO67921.1 | 1.74e-187 | 1 | 388 | 1 | 388 |
| QUT89479.1 | 2.02e-186 | 20 | 388 | 20 | 388 |
| ALJ59478.1 | 4.07e-186 | 20 | 388 | 20 | 388 |
| AUI48832.1 | 1.40e-184 | 20 | 388 | 20 | 388 |
| QCQ31137.1 | 1.98e-184 | 20 | 388 | 20 | 388 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JJA_A | 8.29e-176 | 19 | 388 | 1 | 370 | Crystalstructure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution [Bacteroides fragilis NCTC 9343] |
| 4K05_A | 2.94e-43 | 38 | 386 | 28 | 398 | Crystalstructure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343],4K05_B Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40407 | 4.54e-72 | 33 | 386 | 47 | 414 | Uncharacterized protein YbbC OS=Bacillus subtilis (strain 168) OX=224308 GN=ybbC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
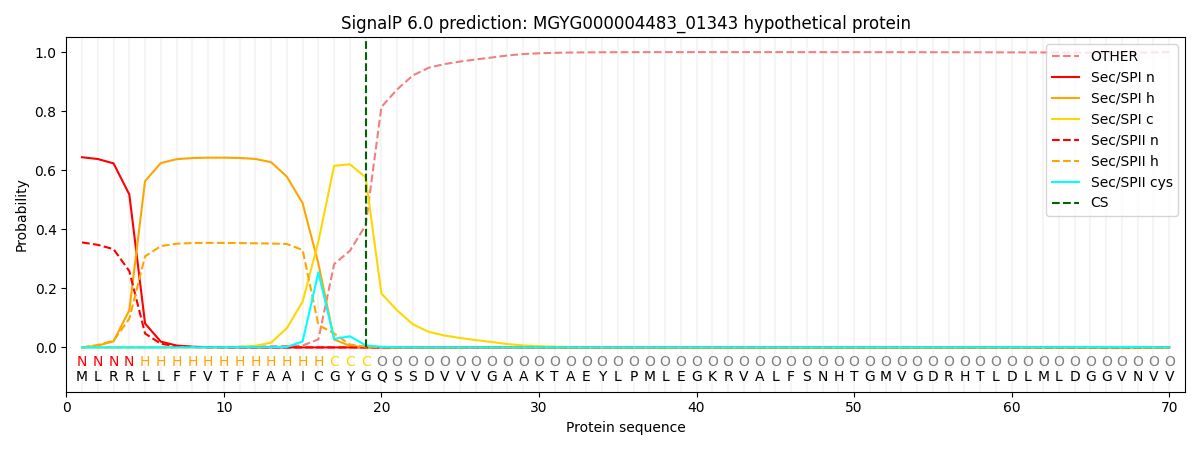
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001292 | 0.634102 | 0.363798 | 0.000332 | 0.000247 | 0.000215 |
