You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004498_00867
You are here: Home > Sequence: MGYG000004498_00867
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-345 sp000433315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RFN20; CAG-288; CAG-345; CAG-345 sp000433315 | |||||||||||
| CAZyme ID | MGYG000004498_00867 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 122715; End: 125354 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.74e-12 | 196 | 462 | 117 | 399 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.41e-06 | 261 | 424 | 88 | 252 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam00754 | F5_F8_type_C | 9.12e-06 | 743 | 867 | 5 | 121 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PLN02793 | PLN02793 | 4.94e-05 | 232 | 344 | 141 | 266 | Probable polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACK41503.1 | 4.32e-106 | 53 | 730 | 25 | 679 |
| ABX43664.1 | 8.95e-106 | 51 | 513 | 35 | 518 |
| ACI19833.1 | 6.55e-104 | 53 | 730 | 25 | 679 |
| QTE67472.1 | 1.43e-98 | 3 | 511 | 1 | 485 |
| BCK00431.1 | 5.72e-96 | 51 | 513 | 57 | 520 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
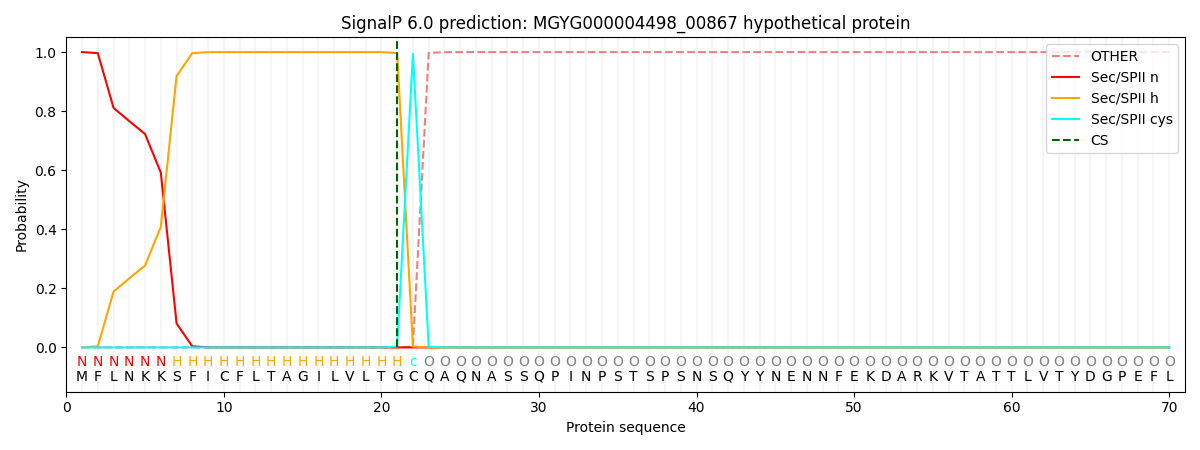
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000066 | 0.000000 | 0.000000 | 0.000000 |
