You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004501_00804
You are here: Home > Sequence: MGYG000004501_00804
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760665 | |||||||||||
| CAZyme ID | MGYG000004501_00804 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6304; End: 9969 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 348 | 887 | 8.6e-139 | 0.6771844660194175 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 32 | 855 | 1 | 868 | alpha-L-rhamnosidase. |
| PRK10150 | PRK10150 | 0.002 | 1064 | 1143 | 56 | 136 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 0.007 | 1066 | 1153 | 58 | 145 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG54195.1 | 1.86e-300 | 26 | 1217 | 35 | 1130 |
| AVM45203.1 | 3.92e-288 | 2 | 1212 | 1 | 1266 |
| AEW21098.1 | 2.81e-285 | 27 | 1217 | 29 | 1121 |
| BAR50760.1 | 8.70e-285 | 27 | 1217 | 32 | 1124 |
| BAR48037.1 | 1.09e-282 | 27 | 1217 | 32 | 1124 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 3.55e-123 | 26 | 1212 | 41 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.55e-110 | 27 | 1219 | 31 | 1105 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 4.05e-110 | 27 | 1219 | 31 | 1105 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 2.00e-171 | 18 | 1215 | 14 | 1158 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
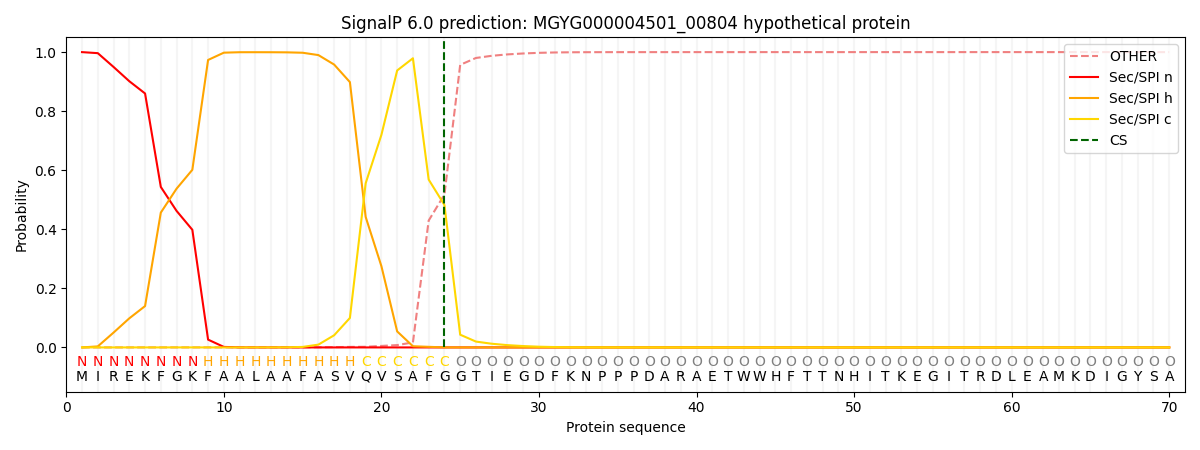
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000574 | 0.998648 | 0.000255 | 0.000182 | 0.000170 | 0.000145 |
