You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004587_01387
You are here: Home > Sequence: MGYG000004587_01387
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1409 sp900553675 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1409; UBA1409 sp900553675 | |||||||||||
| CAZyme ID | MGYG000004587_01387 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16586; End: 17791 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 47 | 228 | 3.2e-70 | 0.9887005649717514 |
| CBM22 | 257 | 383 | 7.3e-30 | 0.9694656488549618 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 1.08e-73 | 47 | 227 | 1 | 175 | Glycosyl hydrolases family 11. |
| pfam02018 | CBM_4_9 | 3.19e-20 | 254 | 387 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL18305.1 | 1.27e-138 | 37 | 399 | 33 | 402 |
| AYF57933.1 | 4.49e-123 | 12 | 398 | 13 | 418 |
| ADD13451.1 | 5.10e-114 | 11 | 399 | 11 | 402 |
| AEE64769.1 | 3.48e-113 | 40 | 395 | 36 | 402 |
| AEE64770.1 | 4.07e-113 | 36 | 395 | 59 | 433 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7AYL_A | 2.06e-92 | 38 | 237 | 32 | 231 | Crystalstructure of the GH11 domain of a multidomain xylanase from the hindgut metagenome of Trinervitermes trinervoides [uncultured bacterium],7AYL_B Crystal structure of the GH11 domain of a multidomain xylanase from the hindgut metagenome of Trinervitermes trinervoides [uncultured bacterium] |
| 6KKA_A | 1.40e-91 | 38 | 237 | 2 | 202 | XylanaseJ mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],6KKA_B Xylanase J mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 6KJL_A | 5.78e-91 | 38 | 237 | 3 | 203 | XylanaseJ from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1],6KJL_B Xylanase J from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1] |
| 2DCJ_A | 1.31e-90 | 38 | 237 | 29 | 229 | Atwo-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCJ_B A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCK_A A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 7AYP_A | 1.10e-89 | 38 | 237 | 4 | 203 | Structureof a GH11 domain refined from the X-ray diffraction data of a GH11-CBM36-1 crystal. [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q53317 | 8.43e-104 | 40 | 399 | 34 | 403 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
| Q8GJ44 | 1.16e-90 | 36 | 244 | 32 | 241 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P17137 | 5.75e-90 | 36 | 236 | 60 | 260 | Endo-1,4-beta-xylanase OS=Clostridium saccharobutylicum OX=169679 GN=xynB PE=3 SV=1 |
| P33558 | 5.04e-88 | 36 | 244 | 32 | 242 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P83513 | 1.97e-86 | 42 | 237 | 22 | 217 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
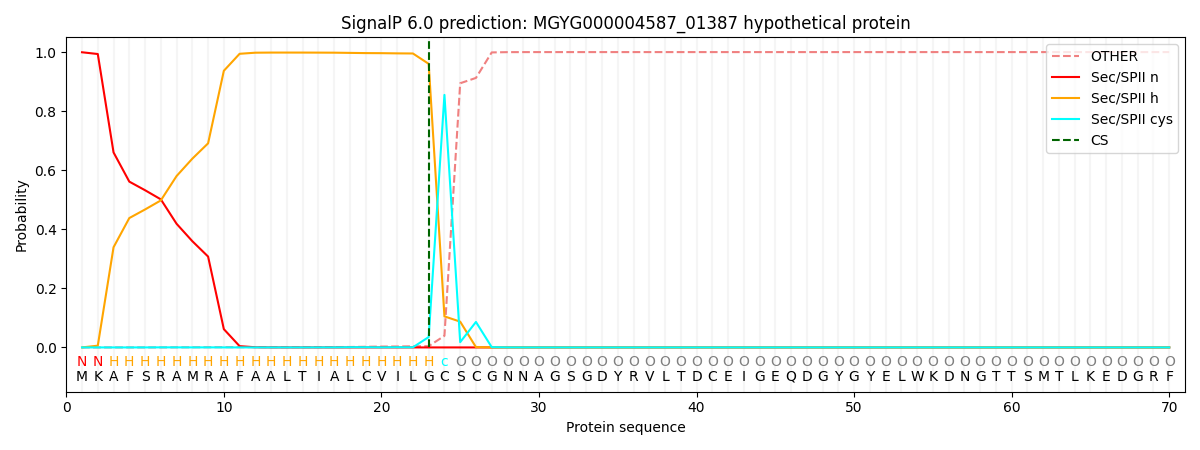
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000002 | 0.000444 | 0.999571 | 0.000000 | 0.000000 | 0.000000 |
