You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004612_01238
You are here: Home > Sequence: MGYG000004612_01238
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
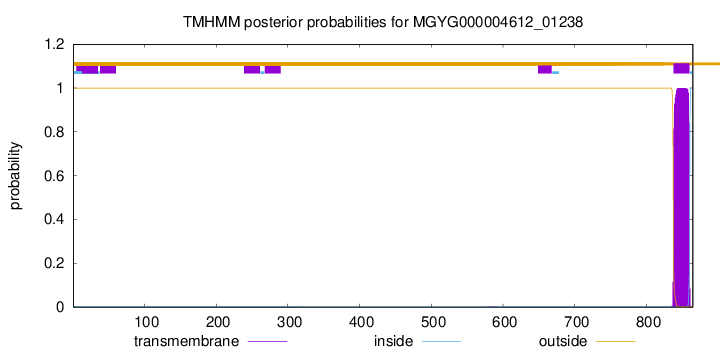
TMHMM annotations
Basic Information help
| Species | UMGS363 sp900768245 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS363; UMGS363 sp900768245 | |||||||||||
| CAZyme ID | MGYG000004612_01238 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 92; End: 2689 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 17 | 678 | 1.6e-201 | 0.9873217115689382 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 4.08e-129 | 19 | 337 | 1 | 293 | Glycosyl hydrolase family 59. |
| COG5520 | XynC | 5.78e-04 | 104 | 288 | 110 | 321 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam17996 | CE2_N | 0.001 | 726 | 793 | 22 | 88 | Carbohydrate esterase 2 N-terminal. This is the N-terminal beta-sheet domain with jelly roll topology found in CE2 acetyl-esterase from the bacterium Clostridium thermocellum. This enzyme displays dual activities, it catalyses the deacetylation of plant polysaccharides and also potentiates the activity of its appended cellulase catalytic module through its noncatalytic cellulose binding function. This N-terminal jelly-roll domain appears to extend the substrate/cellulose binding cleft of the catalytic domain in C.thermocellum. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO04392.1 | 0.0 | 2 | 865 | 42 | 890 |
| CBL16905.1 | 0.0 | 17 | 805 | 1 | 786 |
| CBL34686.1 | 0.0 | 4 | 800 | 21 | 805 |
| CBK96504.1 | 0.0 | 4 | 800 | 21 | 805 |
| BCN31742.1 | 2.31e-293 | 4 | 799 | 8 | 819 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CCC_A | 2.83e-31 | 5 | 604 | 14 | 576 | StructureOf Mouse Galactocerebrosidase With 4nbdg: Enzyme-substrate Complex [Mus musculus],4CCD_A Structure Of Mouse Galactocerebrosidase With D-galactal: Enzyme-intermediate Complex [Mus musculus],4CCE_A Structure Of Mouse Galactocerebrosidase With Galactose: Enzyme-product Complex [Mus musculus],4UFH_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine IGF [Mus musculus],4UFI_A Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF [Mus musculus],4UFJ_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine lactam IGL [Mus musculus],4UFK_A Mouse Galactocerebrosidase complexed with dideoxy-imino-lyxitol DIL [Mus musculus],4UFL_A Mouse Galactocerebrosidase complexed with deoxy-galacto-noeurostegine DGN [Mus musculus],4UFM_A Mouse Galactocerebrosidase complexed with 1-deoxy-galacto-nojirimycin DGJ [Mus musculus],5NXB_A Mouse galactocerebrosidase in complex with saposin A [Mus musculus],5NXB_B Mouse galactocerebrosidase in complex with saposin A [Mus musculus],6Y6S_A Chain A, Galactocerebrosidase [Mus musculus],6Y6T_A Chain A, Galactocerebrosidase [Mus musculus] |
| 3ZR5_A | 2.87e-31 | 5 | 604 | 16 | 578 | STRUCTUREOF GALACTOCEREBROSIDASE FROM MOUSE [Mus musculus],3ZR6_A STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5X3C1 | 5.61e-35 | 23 | 642 | 42 | 628 | Galactocerebrosidase OS=Salmo salar OX=8030 GN=galc PE=2 SV=1 |
| O02791 | 2.02e-34 | 5 | 607 | 44 | 610 | Galactocerebrosidase OS=Macaca mulatta OX=9544 GN=GALC PE=1 SV=2 |
| P54803 | 8.30e-33 | 5 | 607 | 44 | 610 | Galactocerebrosidase OS=Homo sapiens OX=9606 GN=GALC PE=1 SV=3 |
| Q5SNX7 | 6.98e-32 | 23 | 642 | 38 | 623 | Galactocerebrosidase OS=Danio rerio OX=7955 GN=galc PE=2 SV=1 |
| P54804 | 2.31e-31 | 5 | 607 | 28 | 594 | Galactocerebrosidase OS=Canis lupus familiaris OX=9615 GN=GALC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000058 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |