You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004649_00514
You are here: Home > Sequence: MGYG000004649_00514
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Mailhella; | |||||||||||
| CAZyme ID | MGYG000004649_00514 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 258; End: 1787 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 7 | 401 | 7.8e-48 | 0.7351851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.24e-17 | 1 | 385 | 1 | 402 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 4.49e-07 | 33 | 400 | 39 | 424 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARA75637.1 | 1.08e-14 | 1 | 351 | 8 | 365 |
| APS30942.1 | 1.08e-14 | 1 | 351 | 8 | 365 |
| ATV45895.1 | 1.44e-14 | 1 | 351 | 8 | 365 |
| QSD23958.1 | 2.53e-14 | 1 | 351 | 8 | 365 |
| QSD35491.1 | 3.36e-14 | 1 | 351 | 8 | 365 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6DAW3 | 3.52e-13 | 1 | 351 | 8 | 365 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=arnT PE=3 SV=1 |
| Q6D2F3 | 8.19e-13 | 1 | 359 | 8 | 376 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=arnT PE=3 SV=1 |
| B4ETL9 | 9.68e-11 | 28 | 348 | 36 | 365 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Proteus mirabilis (strain HI4320) OX=529507 GN=arnT2 PE=3 SV=1 |
| Q3KCC9 | 1.31e-10 | 35 | 320 | 57 | 353 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1 |
| Q9HY61 | 2.95e-10 | 28 | 351 | 32 | 361 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.987217 | 0.010353 | 0.000943 | 0.000045 | 0.000024 | 0.001420 |
TMHMM Annotations download full data without filtering help
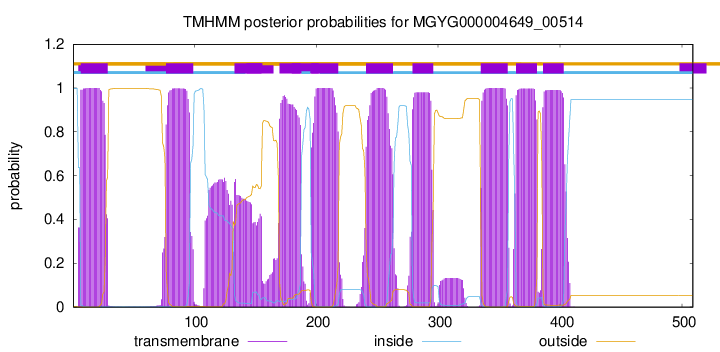
| start | end |
|---|---|
| 7 | 29 |
| 77 | 99 |
| 133 | 155 |
| 170 | 187 |
| 196 | 218 |
| 241 | 263 |
| 279 | 296 |
| 335 | 357 |
| 364 | 381 |
| 386 | 403 |
