You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004688_01391
Basic Information
help
| Species |
CAG-873 sp900550395
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; CAG-873 sp900550395
|
| CAZyme ID |
MGYG000004688_01391
|
| CAZy Family |
GH0 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
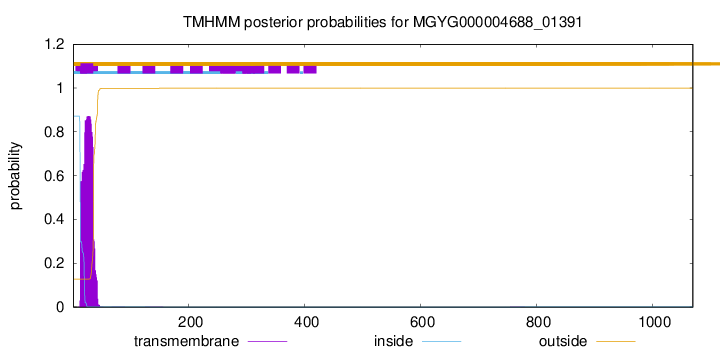
| 1070 |
|
120067.32 |
5.9814 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004688 |
2658373 |
MAG |
Spain |
Europe |
|
| Gene Location |
Start: 29382;
End: 32594
Strand: +
|
No EC number prediction in MGYG000004688_01391.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG3250
|
LacZ |
0.003 |
48 |
213 |
285 |
426 |
Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
This protein is predicted as SP
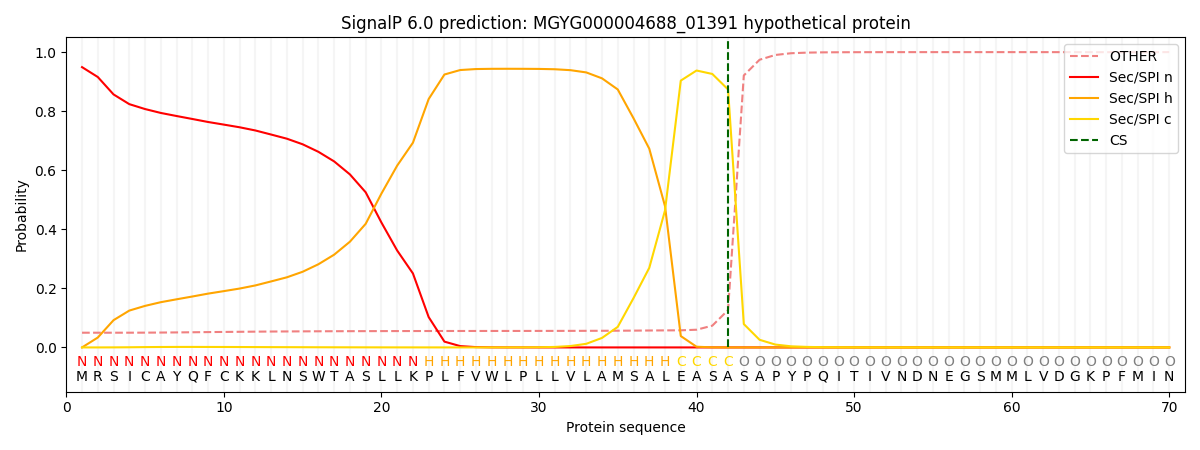
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.052764
|
0.944637
|
0.001751
|
0.000303
|
0.000240
|
0.000263
|