You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004708_01155
You are here: Home > Sequence: MGYG000004708_01155
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000004708_01155 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1016; End: 4117 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 600 | 874 | 8.8e-42 | 0.9236111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 2.07e-15 | 604 | 815 | 89 | 331 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 1.78e-14 | 600 | 830 | 3 | 235 | Pectinesterase. |
| PLN02432 | PLN02432 | 2.99e-14 | 624 | 903 | 40 | 291 | putative pectinesterase |
| PRK10531 | PRK10531 | 2.79e-13 | 634 | 848 | 138 | 381 | putative acyl-CoA thioester hydrolase. |
| PLN02488 | PLN02488 | 3.56e-13 | 600 | 828 | 200 | 430 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH16322.1 | 6.57e-228 | 145 | 1032 | 562 | 1471 |
| AGB28243.1 | 6.52e-226 | 143 | 1031 | 558 | 1369 |
| QUT74167.1 | 3.27e-101 | 9 | 1029 | 262 | 1431 |
| QCD38476.1 | 2.98e-84 | 516 | 1033 | 975 | 1476 |
| QCP72166.1 | 2.98e-84 | 516 | 1033 | 975 | 1476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 3.44e-22 | 600 | 905 | 7 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 2NTB_A | 1.49e-17 | 600 | 905 | 7 | 339 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 1QJV_A | 3.59e-17 | 600 | 905 | 7 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 3UW0_A | 2.06e-15 | 580 | 815 | 12 | 269 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 5C1E_A | 1.95e-13 | 608 | 903 | 24 | 294 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A9 | 1.04e-17 | 576 | 905 | 4 | 363 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| P0C1A8 | 3.36e-17 | 576 | 905 | 4 | 363 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| O04887 | 1.51e-13 | 600 | 905 | 204 | 500 | Pectinesterase 2 OS=Citrus sinensis OX=2711 GN=PECS-2.1 PE=2 SV=1 |
| A2QK82 | 1.47e-12 | 608 | 903 | 52 | 322 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| Q9FM79 | 7.34e-12 | 600 | 830 | 83 | 315 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
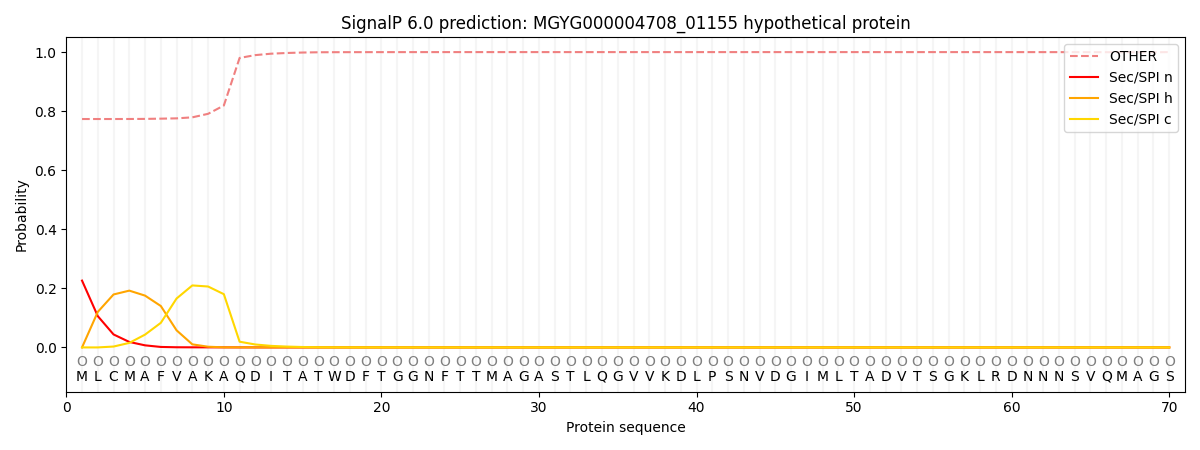
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.780654 | 0.218457 | 0.000550 | 0.000164 | 0.000090 | 0.000107 |
