You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004799_00865
You are here: Home > Sequence: MGYG000004799_00865
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
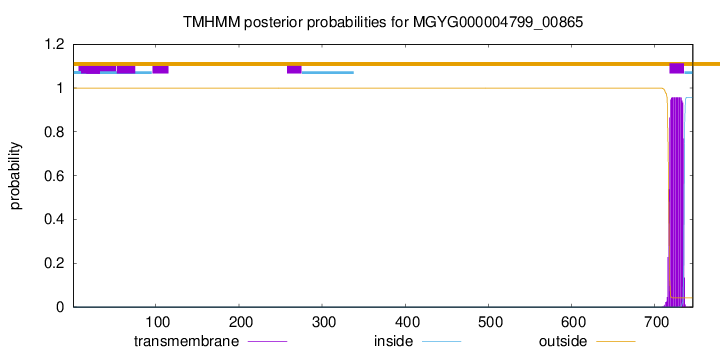
TMHMM annotations
Basic Information help
| Species | Schaedlerella sp900765975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Schaedlerella; Schaedlerella sp900765975 | |||||||||||
| CAZyme ID | MGYG000004799_00865 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16283; End: 18523 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06964 | Alpha-L-AF_C | 9.06e-13 | 406 | 538 | 61 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| COG3534 | AbfA | 1.15e-12 | 2 | 103 | 106 | 211 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 3.74e-12 | 418 | 538 | 67 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| TIGR02243 | TIGR02243 | 1.79e-10 | 98 | 229 | 330 | 442 | putative baseplate assembly protein. This family consists of a large, conserved hypothetical protein in phage tail-like regions of at least six bacterial genomes: Gloeobacter violaceus PCC 7421, Geobacter sulfurreducens PCA, Streptomyces coelicolor A3(2), Streptomyces avermitilis MA-4680, Mesorhizobium loti, and Myxococcus xanthus. The C-terminal region is identified by the broader model pfam04865 as related to baseplate protein J from phage P2, but that relationship is not observed directly. [Mobile and extrachromosomal element functions, Prophage functions] |
| NF033777 | M_group_A_cterm | 1.24e-05 | 551 | 740 | 36 | 218 | M protein C-terminal domain. M protein (emm) is an important virulence protein and serology-defining surface antigen of Streptococcus pyogenes (group A Streptococcus). M protein has an amino-terminal YSIRK-type signal sequence (associated with cross-wall targeting in dividing cells), and a C-terminal LPXTG domain for processing by sortase and covalent attachment to the Gram-positive cell wall. Past the signal peptide, M protein has a hypervariable region, but this HMM describes only the well-conserved region C-terminal to the hypervariable region. It discriminates M protein from two related proteins, Enn and Mrp. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBA48000.1 | 1.35e-197 | 2 | 600 | 428 | 1028 |
| AFL03619.1 | 1.35e-197 | 2 | 600 | 428 | 1028 |
| BAQ97226.1 | 1.90e-197 | 2 | 600 | 428 | 1028 |
| QRI58751.1 | 2.11e-197 | 2 | 600 | 445 | 1045 |
| VEG16797.1 | 2.98e-197 | 2 | 600 | 445 | 1045 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ATW_A | 3.50e-06 | 2 | 76 | 105 | 173 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3S2C_A | 3.51e-06 | 2 | 76 | 105 | 173 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 3UG3_A | 3.61e-06 | 2 | 76 | 125 | 193 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
| 5O7Z_A | 4.73e-06 | 1 | 76 | 104 | 173 | ChainA, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_B Chain B, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_C Chain C, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_D Chain D, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_E Chain E, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O7Z_F Chain F, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_A Chain A, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_B Chain B, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_C Chain C, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_D Chain D, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_E Chain E, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila],5O80_F Chain F, Intracellular exo-alpha-(1->5)-L-arabinofuranosidase [Thermochaetoides thermophila] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
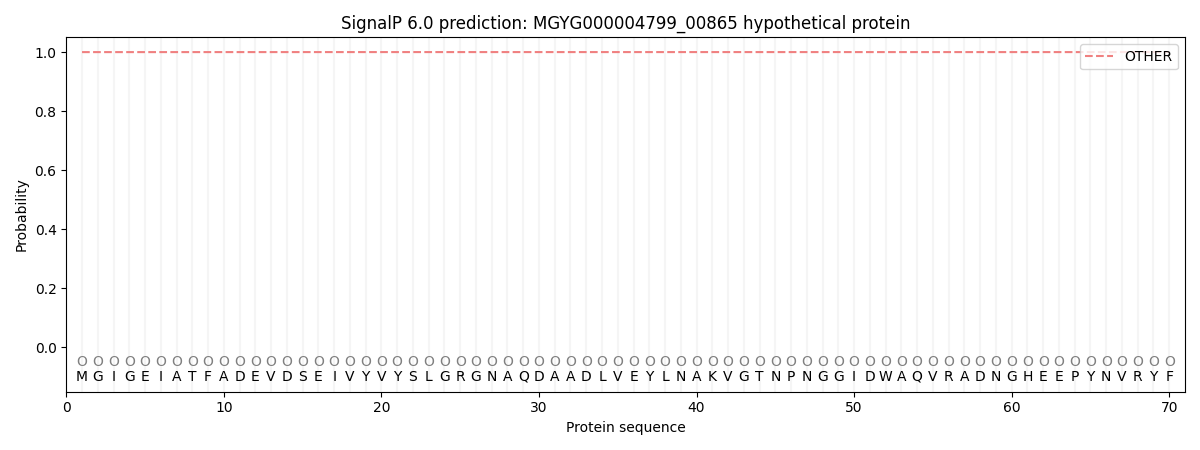
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000041 | 0.000007 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |