You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004872_01566
You are here: Home > Sequence: MGYG000004872_01566
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
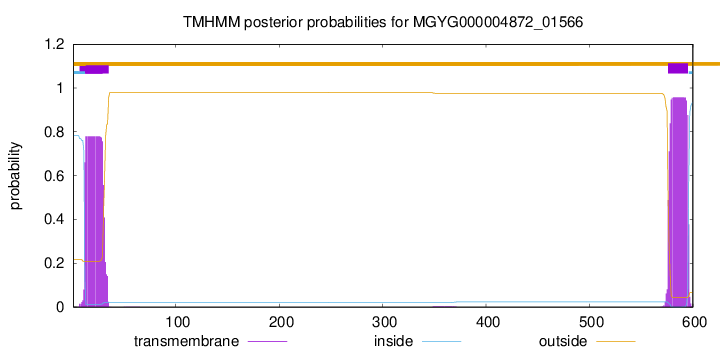
TMHMM annotations
Basic Information help
| Species | Stoquefichus sp001244545 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Stoquefichus; Stoquefichus sp001244545 | |||||||||||
| CAZyme ID | MGYG000004872_01566 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43775; End: 45577 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11790 | Glyco_hydro_cc | 3.85e-45 | 134 | 391 | 1 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
| pfam09479 | Flg_new | 2.12e-10 | 496 | 549 | 10 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| NF033189 | internalin_A | 3.40e-09 | 433 | 552 | 582 | 704 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| NF033189 | internalin_A | 1.28e-08 | 428 | 550 | 507 | 632 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| NF033188 | internalin_H | 1.36e-08 | 426 | 552 | 351 | 482 | InlH/InlC2 family class 1 internalin. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin H (InlH), or internalin C2, two class 1 (LPXTG-type) internalins that are closely related, one apparently derived from the other through a recombination event. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQV06004.1 | 1.16e-134 | 4 | 575 | 4 | 575 |
| QQY27035.1 | 9.18e-134 | 4 | 575 | 4 | 575 |
| QPS14141.1 | 5.13e-133 | 4 | 575 | 4 | 575 |
| QMW75524.1 | 5.13e-133 | 4 | 575 | 4 | 575 |
| EDS18726.1 | 5.13e-133 | 4 | 575 | 4 | 575 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UAQ_A | 2.26e-28 | 129 | 362 | 24 | 245 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
| 6UFL_A | 5.74e-28 | 129 | 362 | 24 | 245 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
| 6UAS_A | 1.45e-27 | 129 | 362 | 24 | 245 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei] |
| 6UAT_A | 1.45e-27 | 129 | 362 | 24 | 245 | Crystalstructure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei],6UAU_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose [Amycolatopsis mediterranei] |
| 6UBC_A | 1.35e-10 | 130 | 391 | 203 | 436 | ChainA, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DJ98 | 1.95e-13 | 432 | 550 | 99 | 220 | Putative membrane protein Bcell_0381 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0381 PE=4 SV=1 |
| Q723K6 | 2.90e-07 | 428 | 550 | 508 | 633 | Internalin A OS=Listeria monocytogenes serotype 4b (strain F2365) OX=265669 GN=inlA PE=3 SV=1 |
| G2K3G6 | 3.82e-07 | 433 | 552 | 583 | 705 | Internalin A OS=Listeria monocytogenes serotype 1/2a (strain 10403S) OX=393133 GN=inlA PE=3 SV=1 |
| P0DJM0 | 5.03e-07 | 433 | 552 | 583 | 705 | Internalin A OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlA PE=1 SV=1 |
| Q7AP87 | 8.79e-06 | 438 | 552 | 365 | 482 | Internalin H OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
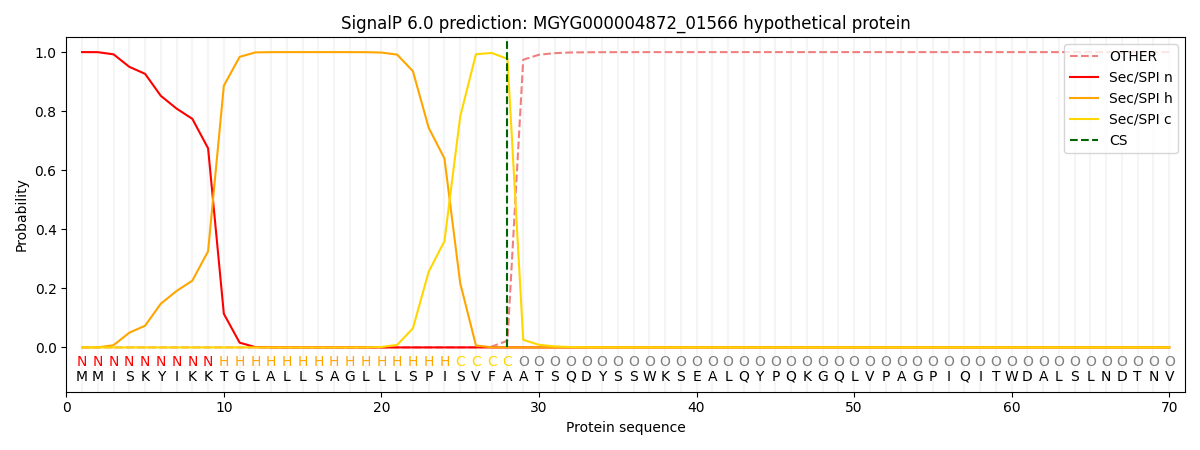
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000363 | 0.998857 | 0.000197 | 0.000225 | 0.000179 | 0.000161 |