You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004891_02574
You are here: Home > Sequence: MGYG000004891_02574
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; KM106-2; | |||||||||||
| CAZyme ID | MGYG000004891_02574 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17999; End: 20032 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 132 | 350 | 7e-40 | 0.7772277227722773 |
| CBM77 | 570 | 672 | 5.8e-35 | 0.9805825242718447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18283 | CBM77 | 1.26e-34 | 568 | 674 | 2 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| smart00656 | Amb_all | 5.99e-24 | 197 | 339 | 58 | 160 | Amb_all domain. |
| COG3866 | PelB | 4.60e-19 | 36 | 345 | 20 | 251 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 2.29e-12 | 198 | 343 | 78 | 189 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| pfam04886 | PT | 1.43e-11 | 503 | 537 | 1 | 35 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEH70392.1 | 1.38e-174 | 48 | 482 | 1326 | 1758 |
| ACZ98653.1 | 6.37e-160 | 48 | 482 | 913 | 1353 |
| ADD62022.1 | 2.47e-131 | 51 | 483 | 1600 | 2035 |
| QJS18636.1 | 2.25e-96 | 48 | 477 | 245 | 673 |
| ANF98673.1 | 4.00e-88 | 48 | 414 | 21 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 1.03e-83 | 45 | 415 | 4 | 377 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
| 5FU5_A | 1.17e-18 | 571 | 674 | 10 | 111 | Thecomplexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition [Ruminococcus flavefaciens] |
| 3ZSC_A | 7.99e-18 | 143 | 339 | 63 | 210 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 1VBL_A | 1.60e-12 | 158 | 347 | 143 | 306 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 5AMV_A | 3.36e-11 | 169 | 345 | 148 | 298 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 8.08e-84 | 45 | 381 | 33 | 368 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
| Q9WYR4 | 4.24e-18 | 143 | 339 | 90 | 237 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
| B1L969 | 1.34e-17 | 143 | 339 | 88 | 235 | Pectate trisaccharide-lyase OS=Thermotoga sp. (strain RQ2) OX=126740 GN=pelA PE=3 SV=1 |
| Q9FM66 | 1.78e-10 | 164 | 341 | 154 | 284 | Putative pectate lyase 21 OS=Arabidopsis thaliana OX=3702 GN=At5g55720 PE=3 SV=1 |
| P39116 | 2.01e-10 | 169 | 345 | 169 | 319 | Pectate lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pel PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
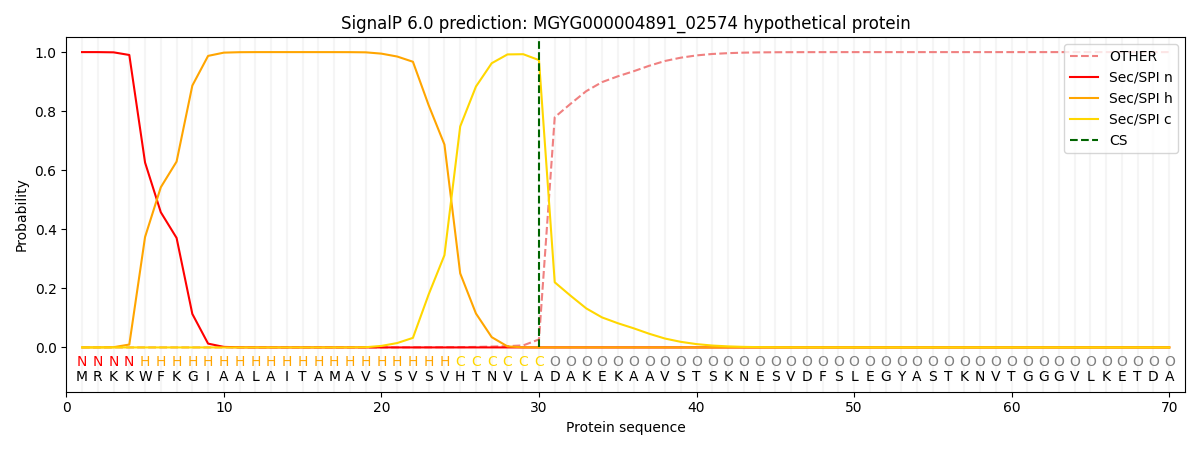
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000310 | 0.998917 | 0.000253 | 0.000179 | 0.000169 | 0.000148 |
